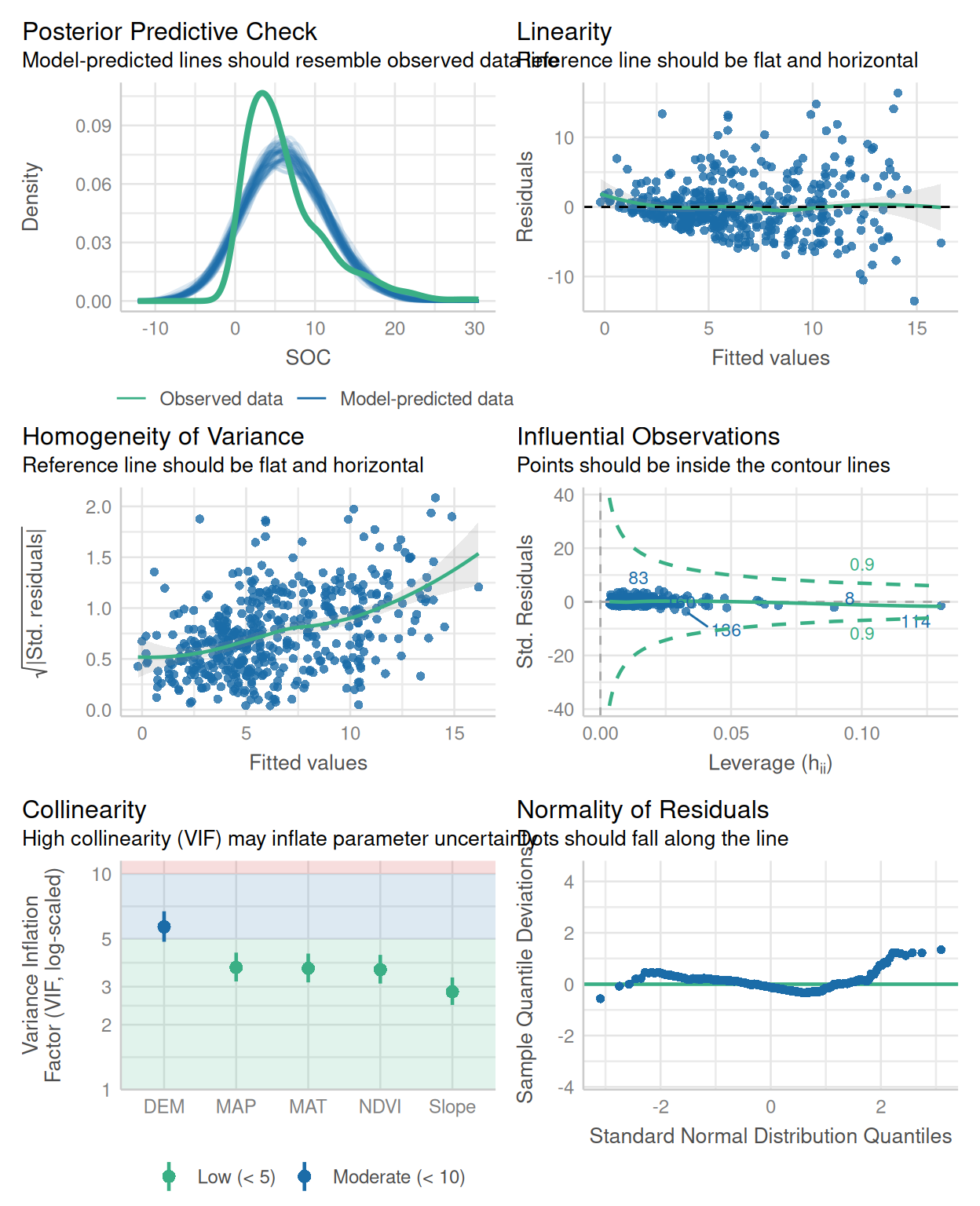
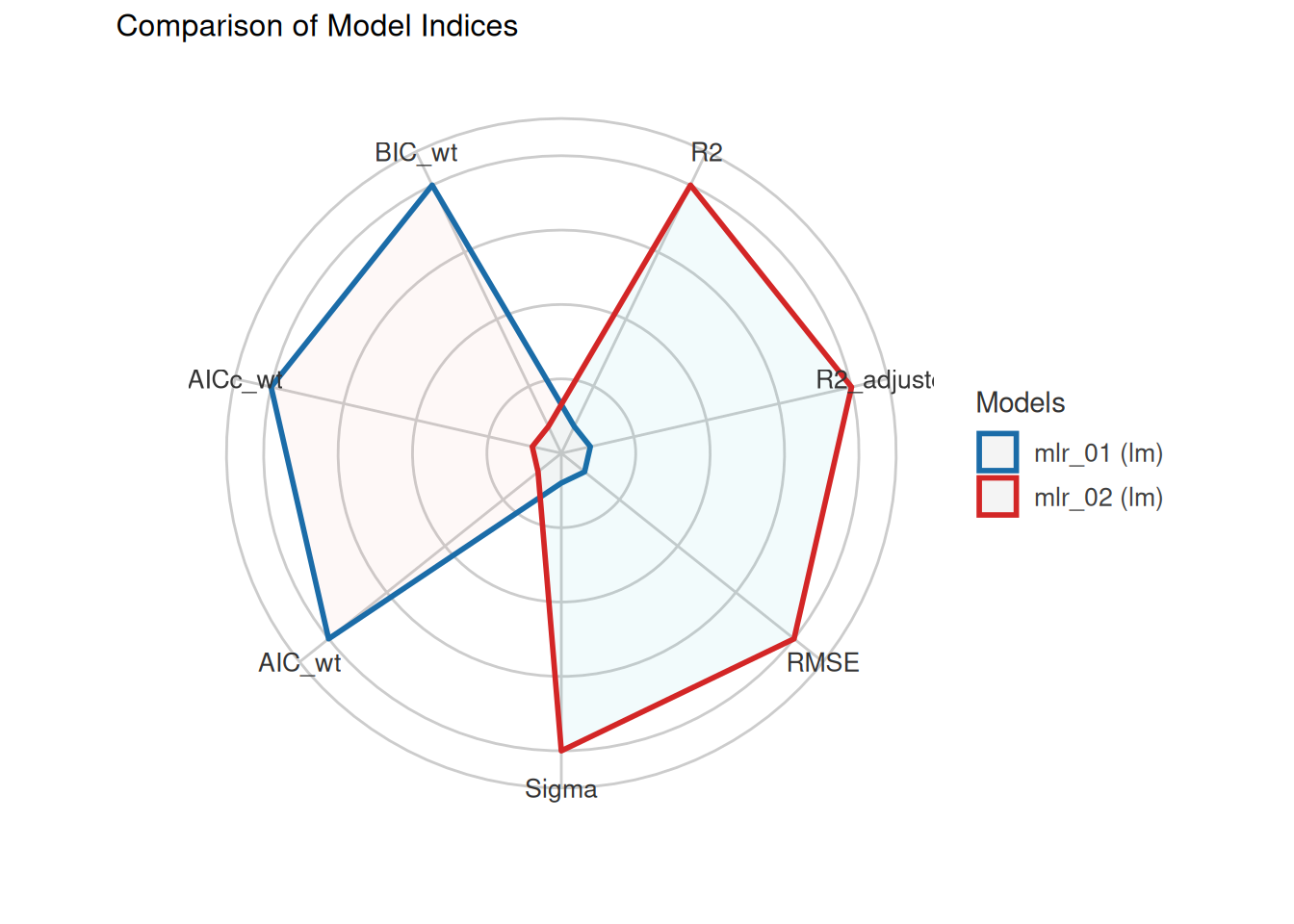
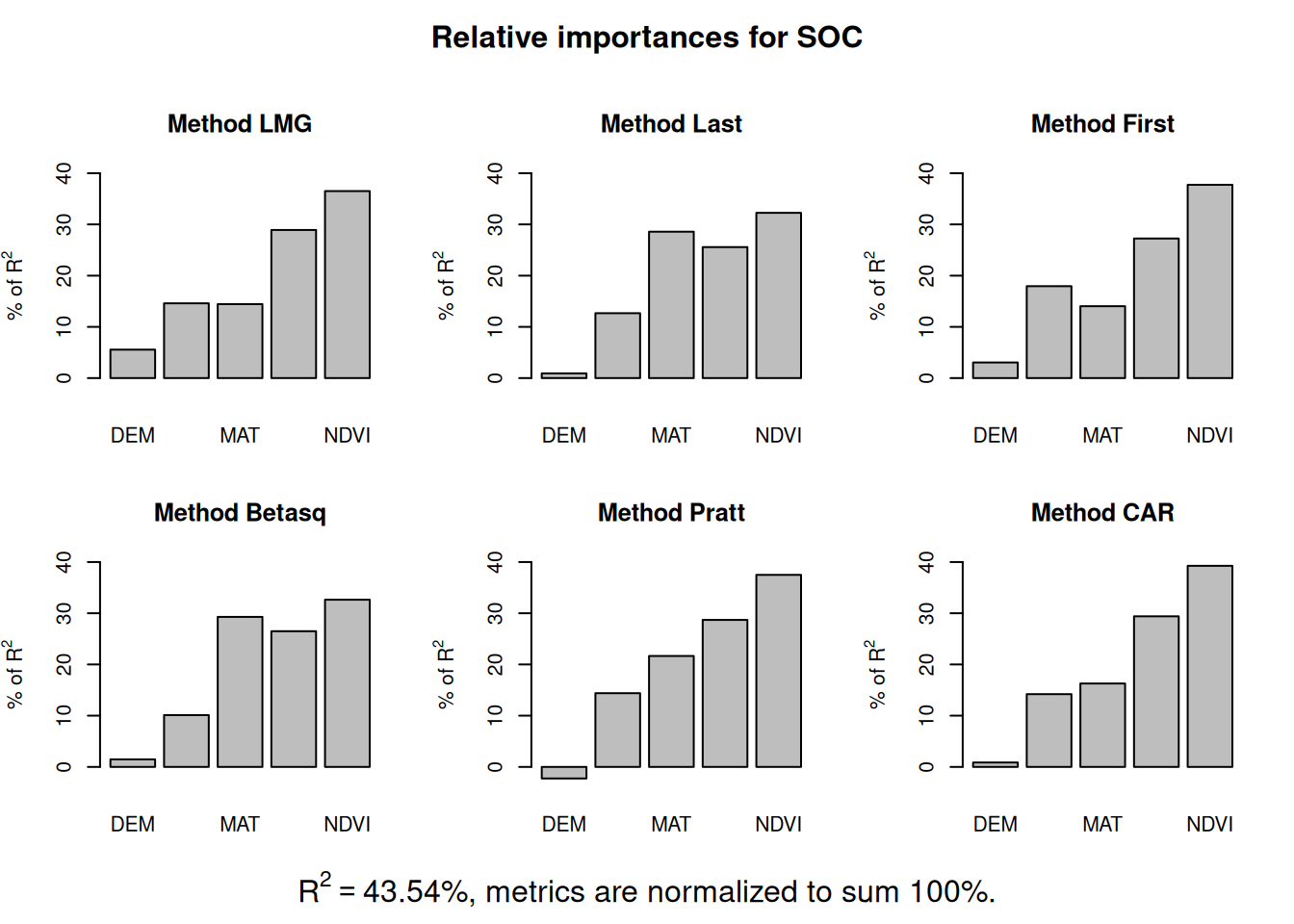
We fitted a linear model (estimated using OLS) to predict SOC with DEM, Slope,
MAT, MAP, NDVI, NLCD and FRG (formula: SOC ~ DEM + Slope + MAT + MAP + NDVI +
NLCD + FRG). The model explains a statistically significant and substantial
proportion of variance (R2 = 0.45, F(13, 453) = 28.67, p < .001, adj. R2 =
0.44). The model's intercept, corresponding to DEM = 0, Slope = 0, MAT = 0, MAP
= 0, NDVI = 0, NLCD = Forest and FRG = Fire Regime Group I, is at 2.53 (95% CI
[-3.04, 8.09], t(453) = 0.89, p = 0.372). Within this model:
- The effect of DEM is statistically non-significant and negative (beta =
-4.48e-04, 95% CI [-1.79e-03, 8.92e-04], t(453) = -0.66, p = 0.511; Std. beta =
-0.07, 95% CI [-0.27, 0.14])
- The effect of Slope is statistically significant and positive (beta = 0.16,
95% CI [0.02, 0.29], t(453) = 2.27, p = 0.024; Std. beta = 0.14, 95% CI [0.02,
0.27])
- The effect of MAT is statistically significant and negative (beta = -0.40,
95% CI [-0.60, -0.20], t(453) = -3.98, p < .001; Std. beta = -0.33, 95% CI
[-0.49, -0.16])
- The effect of MAP is statistically significant and positive (beta = 6.76e-03,
95% CI [3.26e-03, 0.01], t(453) = 3.80, p < .001; Std. beta = 0.28, 95% CI
[0.13, 0.42])
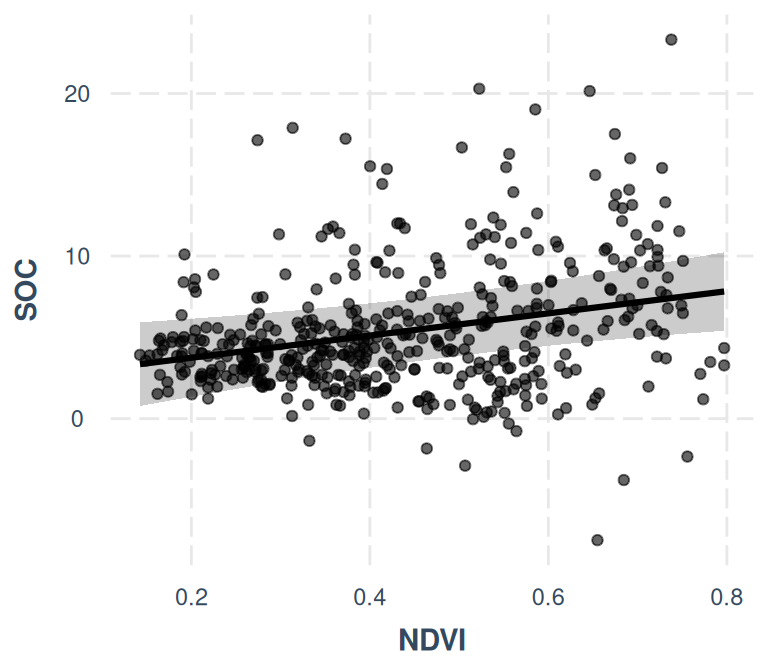
- The effect of NDVI is statistically significant and positive (beta = 6.83,
95% CI [2.01, 11.65], t(453) = 2.78, p = 0.006; Std. beta = 0.22, 95% CI [0.06,
0.37])
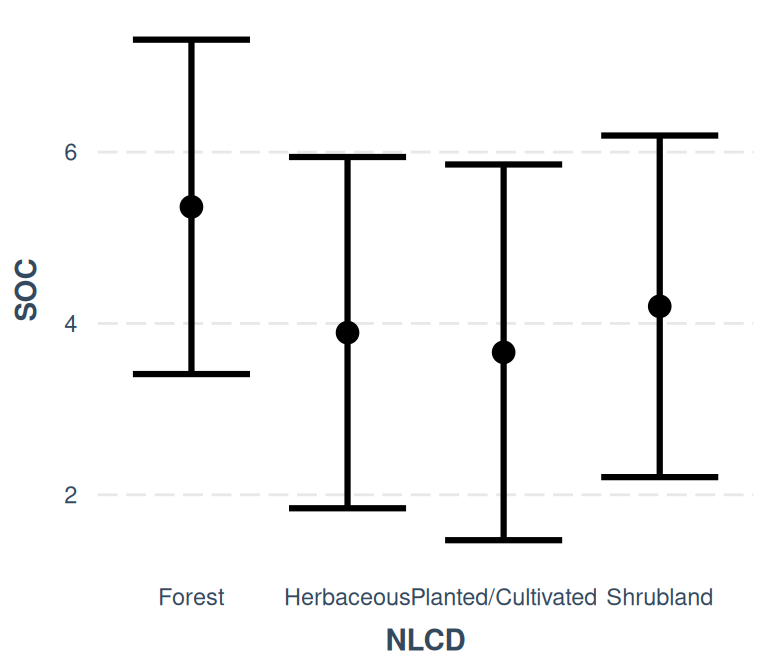
- The effect of NLCD [Herbaceous] is statistically non-significant and negative
(beta = -1.47, 95% CI [-3.20, 0.26], t(453) = -1.67, p = 0.096; Std. beta =
-0.29, 95% CI [-0.63, 0.05])
- The effect of NLCD [Planted/Cultivated] is statistically non-significant and
negative (beta = -1.70, 95% CI [-3.64, 0.25], t(453) = -1.72, p = 0.087; Std.
beta = -0.34, 95% CI [-0.72, 0.05])
- The effect of NLCD [Shrubland] is statistically non-significant and negative
(beta = -1.16, 95% CI [-2.73, 0.41], t(453) = -1.46, p = 0.146; Std. beta =
-0.23, 95% CI [-0.54, 0.08])
- The effect of FRG [Fire Regime Group II] is statistically significant and
positive (beta = 2.90, 95% CI [0.86, 4.94], t(453) = 2.79, p = 0.005; Std. beta
= 0.57, 95% CI [0.17, 0.98])
- The effect of FRG [Fire Regime Group III] is statistically non-significant
and positive (beta = 1.58, 95% CI [-0.39, 3.55], t(453) = 1.58, p = 0.115; Std.
beta = 0.31, 95% CI [-0.08, 0.70])
- The effect of FRG [Fire Regime Group IV] is statistically non-significant and
positive (beta = 1.13, 95% CI [-1.10, 3.36], t(453) = 1.00, p = 0.319; Std.
beta = 0.22, 95% CI [-0.22, 0.67])
- The effect of FRG [Fire Regime Group V] is statistically non-significant and
positive (beta = 0.96, 95% CI [-1.78, 3.69], t(453) = 0.69, p = 0.492; Std.
beta = 0.19, 95% CI [-0.35, 0.73])
- The effect of FRG [Indeterminate FRG] is statistically non-significant and
positive (beta = 1.79, 95% CI [-1.63, 5.21], t(453) = 1.03, p = 0.304; Std.
beta = 0.35, 95% CI [-0.32, 1.03])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald t-distribution approximation.