Code
packages <- c('tidyverse',
'dlookr',
'flextable'
)This notebook is a tutorial on how to use the {dlookr} package for exploratory data analysis (EDA) in R. The {dlookr} package provides a comprehensive set of tools for diagnosing, exploring, and transforming data. It includes functions for data quality diagnosis, descriptive statistics, correlation analysis, and data transformation.
The {dlookr} is a collection of tools that support data diagnosis, exploration, and transformation. Data diagnostics provides information and visualization of missing values and outliers and unique and negative values to help you understand the distribution and quality of your data. Data exploration provides information and visualization of the descriptive statistics of univariate variables, normality tests and outliers, correlation of two variables, and relationship between target variable and predictor. Data transformation supports binning for categorizing continuous variables, imputates missing values and outliers, resolving skewness. And it creates automated reports that support these three tasks.

Features:
Diagnose data quality.
Find appropriate scenarios to pursuit the follow-up analysis through data exploration and understanding.
Derive new variables or perform variable transformations.
Automatically generate reports for the above three tasks.
Supports quality diagnosis and EDA of table of DBMS
Data quality diagnosis for data.frame, tbl_df, and table of DBMS
Exploratory Data Analysis for data.frame, tbl_df, and table of DBMS
Data Transformation
Data diagnosis and EDA for table of DBMS
install.packages(c("nloptr", "lme4", "jomo", "mitml", 'mice', 'devtools'))
devtools::install_github("choonghyunryu/dlookr")#| warning: false
#| error: false
# Install missing packages
new_packages <- packages[!(packages %in% installed.packages()[,"Package"])]
if(length(new_packages)) install.packages(new_packages)
Successfully loaded packages: [1] "package:flextable" "package:dlookr" "package:lubridate"
[4] "package:forcats" "package:stringr" "package:dplyr"
[7] "package:purrr" "package:readr" "package:tidyr"
[10] "package:tibble" "package:ggplot2" "package:tidyverse"
[13] "package:stats" "package:graphics" "package:grDevices"
[16] "package:utils" "package:datasets" "package:methods"
[19] "package:base" The data set use in this exercise can be downloaded from my Dropbox or from my Github account.
We will use read_csv() function of readr package to import data as a tidy data.
Data Quality Diagnosis is the first step before any statistical analysis. We use diagnose() function of dlookr package to do general General diagnosis of all variables.
The variables of the tbl_df object returned by diagnose () are as follows:
variables : variable names
types : the data type of the variables
missing_count : number of missing values
missing_percent : percentage of missing values
unique_count : number of unique values
unique_rate : rate of unique value. unique_count / number of observation
variables | types | missing_count | missing_percent | unique_count | unique_rate |
|---|---|---|---|---|---|
ID | numeric | 0 | 0.0000000 | 471 | 1.000000000 |
FIPS | numeric | 0 | 0.0000000 | 172 | 0.365180467 |
STATE_ID | numeric | 0 | 0.0000000 | 4 | 0.008492569 |
STATE | character | 0 | 0.0000000 | 4 | 0.008492569 |
COUNTY | character | 0 | 0.0000000 | 161 | 0.341825902 |
Longitude | numeric | 0 | 0.0000000 | 471 | 1.000000000 |
Latitude | numeric | 0 | 0.0000000 | 471 | 1.000000000 |
SOC | numeric | 4 | 0.8492569 | 457 | 0.970276008 |
DEM | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
Aspect | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
Slope | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
TPI | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
KFactor | numeric | 0 | 0.0000000 | 386 | 0.819532909 |
MAP | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
MAT | numeric | 0 | 0.0000000 | 463 | 0.983014862 |
NDVI | numeric | 0 | 0.0000000 | 464 | 0.985138004 |
SiltClay | numeric | 0 | 0.0000000 | 462 | 0.980891720 |
NLCD | character | 0 | 0.0000000 | 4 | 0.008492569 |
FRG | character | 0 | 0.0000000 | 6 | 0.012738854 |
Missing Value(NA) : Variables with many missing values, i.e. those with a missing_percent close to 100, should be excluded from the analysis.
Unique value : Variables with a unique value (unique_count = 1) are considered to be excluded from data analysis. And if the data type is not numeric (integer, numeric) and the number of unique values is equal to the number of observations (unique_rate = 1), then the variable is likely to be an identifier. Therefore, this variable is also not suitable for the analysis model.
We may use diagnose_numeric(), diagnoses numeric(continuous and discrete) variables in a data frame returns more diagnostic information such as:
min : minimum value
Q1 : 1/4 quartile, 25th percentile
mean : arithmetic mean
median : median, 50th percentile
Q3 : 3/4 quartile, 75th percentile
max : maximum value
zero : number of observations with a value of 0
minus : number of observations with negative numbers
outlier : number of outliers
variables | min | Q1 | mean | median | Q3 | max | zero | minus | outlier |
|---|---|---|---|---|---|---|---|---|---|
SOC | 0.4080000 | 2.7695000 | 6.3507623126 | 4.97100000 | 8.7135000 | 30.4730000 | 0 | 0 | 20 |
DEM | 258.6488037 | 1,175.3313595 | 1,631.1063060667 | 1,592.89318800 | 2,234.2648930 | 3,618.0241700 | 0 | 0 | 0 |
Slope | 0.6492527 | 1.4506671 | 4.8267398902 | 2.72667742 | 7.1070788 | 26.1041622 | 0 | 0 | 20 |
Aspect | 86.8945694 | 148.8052292 | 165.4676589153 | 164.07072450 | 179.0842895 | 255.8335266 | 0 | 0 | 8 |
TPI | -26.7086506 | -0.8160543 | -0.0006690991 | -0.04758827 | 0.8490718 | 16.7062569 | 0 | 241 | 85 |
KFactor | 0.0500000 | 0.1933357 | 0.2558965090 | 0.28000000 | 0.3200000 | 0.4300000 | 0 | 0 | 0 |
MAP | 193.9132233 | 352.7745056 | 499.3729530231 | 432.63040160 | 590.4269104 | 1,128.1145020 | 0 | 0 | 17 |
MAT | -0.5910638 | 5.8800533 | 8.8855210982 | 9.17283535 | 12.4442859 | 16.8742866 | 0 | 6 | 0 |
NDVI | 0.1424335 | 0.3053468 | 0.4354311144 | 0.41568252 | 0.5559025 | 0.7969922 | 0 | 0 | 0 |
SiltClay | 9.1619568 | 42.7587299 | 53.6779156034 | 52.11276627 | 62.8508625 | 89.8344116 | 0 | 0 | 3 |
diagnose_category() diagnoses the categorical(factor, ordered, character) variables of a data frame. The usage is similar to diagnose() but returns more diagnostic information such as:
variables : variable names
levels: level names
N : number of observation
freq : number of observation at the levels
ratio : percentage of observation at the levels
rank : rank of occupancy ratio of levels
variables | levels | N | freq | ratio | rank |
|---|---|---|---|---|---|
STATE | Colorado | 471 | 136 | 28.874735 | 1 |
STATE | Wyoming | 471 | 120 | 25.477707 | 2 |
STATE | New Mexico | 471 | 109 | 23.142251 | 3 |
STATE | Kansas | 471 | 106 | 22.505308 | 4 |
NLCD | Herbaceous | 471 | 151 | 32.059448 | 1 |
NLCD | Shrubland | 471 | 130 | 27.600849 | 2 |
NLCD | Planted/Cultivated | 471 | 97 | 20.594480 | 3 |
NLCD | Forest | 471 | 93 | 19.745223 | 4 |
FRG | Fire Regime Group II | 471 | 252 | 53.503185 | 1 |
FRG | Fire Regime Group III | 471 | 100 | 21.231423 | 2 |
FRG | Fire Regime Group IV | 471 | 75 | 15.923567 | 3 |
FRG | Fire Regime Group I | 471 | 19 | 4.033970 | 4 |
FRG | Fire Regime Group V | 471 | 18 | 3.821656 | 5 |
FRG | Indeterminate FRG | 471 | 7 | 1.486200 | 6 |
diagnose_outlier() diagnoses the outliers of the numeric (continuous and discrete) variables of the data frame.
outliers_cnt : number of outliers
outliers_ratio : percent of outliers
outliers_mean : arithmetic average of outliers
with_mean : arithmetic average of with outliers
without_mean : arithmetic average of without outliers
The diagnose_outlier() produces outlier information for diagnosing the quality of the numerical data.
# A tibble: 10 × 6
variables outliers_cnt outliers_ratio outliers_mean with_mean without_mean
<chr> <int> <dbl> <dbl> <dbl> <dbl>
1 SOC 20 4.25 21.1 6.35 5.69
2 DEM 0 0 NaN 1631. 1631.
3 Slope 20 4.25 18.9 4.83 4.20
4 Aspect 8 1.70 224. 165. 164.
5 TPI 85 18.0 0.291 -0.000669 -0.0649
6 KFactor 0 0 NaN 0.256 0.256
7 MAP 17 3.61 1049. 499. 479.
8 MAT 0 0 NaN 8.89 8.89
9 NDVI 0 0 NaN 0.435 0.435
10 SiltClay 3 0.637 9.71 53.7 54.0 plot_outlier() visualizes outliers of numerical variables(continuous and discrete) of data.frame. Usage is the same diagnose().
The plot derived from the numerical data diagnosis is as follows.
With outliers box plot
Without outliers box plot
With outliers histogram
Without outliers histogram
The following example uses plot_outlier() after diagnose_outlier(), and filter and select functions with dplyr packages to visualize this with an outlier ratio of 0.5% or higher.
normality() function of dlookr performs a normality test on multiple numerical data. Shapiro-Wilk normality test is performed. When the number of observations is greater than 5000, it is tested after extracting 5000 samples by random simple sampling.
The variables of tbl_df object returned by normality() are as follows.
statistic : Statistics of the Shapiro-Wilk test
p_value : p-value of the Shapiro-Wilk test
sample : Number of sample observations performed Shapiro-Wilk test
vars | statistic | p_value | sample |
|---|---|---|---|
SOC | 0.8723726 | 0.0000000000000000003914388 | 471 |
MAP | 0.8970027 | 0.0000000000000000287526353 | 471 |
NDVI | 0.9698609 | 0.0000000291124642567419318 | 471 |
DEM | 0.9731601 | 0.0000001328862497056715322 | 471 |
MAT | 0.9732952 | 0.0000001417229160828437308 | 471 |
The normality() function supports the group_by() function syntax in the dplyr package.
variable | NLCD | statistic | p_value | sample |
|---|---|---|---|---|
SOC | Planted/Cultivated | 0.9693901 | 0.02290858185622282 | 97 |
SOC | Forest | 0.9264632 | 0.00005866058197987 | 93 |
SOC | Herbaceous | 0.8892600 | 0.00000000342072181 | 151 |
SOC | Shrubland | 0.8207045 | 0.00000000003809702 | 130 |
We may also use plot_normality() function of dlookr package to visualizes the normality of numeric data. The information that plot_normality() visualizes is as follows.
Histogram of original data
Q-Q plot of original data
histogram of log transformed data
Histogram of square root transformed data
The describe() function from dloookr package computes descriptive statistics for numerical data. The descriptive statistics help determine the distribution of numerical variables.
The variables of the tbl_df object returned by describe() are as follows.
n : number of observations excluding missing values
na : number of missing values
mean : arithmetic average
sd : standard deviation
se_mean : standard error mean. sd/sqrt(n)
IQR : interquartile range (Q3-Q1)
skewness : skewness
kurtosis : kurtosis
p25 : Q1. 25% percentile
p50 : median. 50% percentile
p75 : Q3. 75% percentile
p01, p05, p10, p20, p30 : 1%, 5%, 20%, 30% percentiles
p40, p60, p70, p80 : 40%, 60%, 70%, 80% percentiles
p90, p95, p99, p100 : 90%, 95%, 99%, 100% percentiles
described_variables | n | na | mean | sd | se_mean | IQR | skewness | kurtosis | p00 | p01 | p05 | p10 | p20 | p25 | p30 | p40 | p50 | p60 | p70 | p75 | p80 | p90 | p95 | p99 | p100 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SOC | 467 | 4 | 6.3507623 | 5.0454091 | 0.233473691 | 5.9440000 | 1.46472837 | 2.4271923 | 0.4080000 | 0.4909400 | 0.9637000 | 1.2902000 | 2.3294000 | 2.7695000 | 3.1114000 | 3.9906000 | 4.9710000 | 6.1266000 | 7.5030000 | 8.7135000 | 10.0522000 | 13.3830000 | 16.5219000 | 22.1247600 | 30.4730000 |
DEM | 471 | 0 | 1,631.1063061 | 767.6923254 | 35.373395140 | 1,058.9335335 | -0.02350235 | -0.8039161 | 258.6488037 | 288.9806610 | 353.7696533 | 441.8609924 | 925.0328369 | 1,175.3313595 | 1,271.5794680 | 1,400.3239750 | 1,592.8931880 | 1,876.8692630 | 2,164.8010250 | 2,234.2648930 | 2,334.0241700 | 2,620.1455080 | 2,797.0867920 | 3,157.0538086 | 3,618.0241700 |
MAP | 471 | 0 | 499.3729530 | 206.9359198 | 9.535103866 | 237.6524048 | 1.08253930 | 0.4698226 | 193.9132233 | 205.0283020 | 261.5091248 | 290.6307068 | 340.8447266 | 352.7745056 | 371.9525452 | 404.3808899 | 432.6304016 | 471.3896484 | 557.4978027 | 590.4269104 | 663.0267944 | 835.6693726 | 927.7701416 | 1,102.3618041 | 1,128.1145020 |
MAT | 471 | 0 | 8.8855211 | 4.0981336 | 0.188832030 | 6.5642326 | -0.27522458 | -0.8236567 | -0.5910638 | -0.1158469 | 1.6258606 | 2.9455154 | 5.0193229 | 5.8800533 | 6.8264499 | 7.5748353 | 9.1728353 | 10.5665998 | 11.8026342 | 12.4442859 | 12.7931766 | 13.9399471 | 14.6389923 | 16.2291578 | 16.8742866 |
NDVI | 471 | 0 | 0.4354311 | 0.1620239 | 0.007465669 | 0.2505557 | 0.23375088 | -0.9180418 | 0.1424335 | 0.1631678 | 0.1920738 | 0.2215059 | 0.2756113 | 0.3053468 | 0.3317074 | 0.3769788 | 0.4156825 | 0.4773465 | 0.5348566 | 0.5559025 | 0.5853671 | 0.6759516 | 0.7216224 | 0.7601239 | 0.7969922 |
The describe() function supports the group_by() function syntax of the dplyr package. Following function calculate descriptive testatrices of SOC and NDVI of different NLCD
described_variables | NLCD | n | na | mean | sd | se_mean | IQR | skewness | kurtosis | p00 | p01 | p05 | p10 | p20 | p25 | p30 | p40 | p50 | p60 | p70 | p75 | p80 | p90 | p95 | p99 | p100 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
NDVI | Forest | 93 | 0 | 0.5705648 | 0.1155016 | 0.01197696 | 0.1165678 | -0.6719584 | 0.1157272 | 0.2830779 | 0.2856635 | 0.3427034 | 0.3610759 | 0.5045352 | 0.5326702 | 0.5390721 | 0.5576768 | 0.5759758 | 0.6085463 | 0.6290170 | 0.6492380 | 0.6708930 | 0.7010803 | 0.7353695 | 0.7711932 | 0.7814745 |
NDVI | Herbaceous | 151 | 0 | 0.4003131 | 0.1307054 | 0.01063666 | 0.1257634 | 0.9764992 | 0.4084170 | 0.1648289 | 0.1785699 | 0.2555612 | 0.2651424 | 0.2944161 | 0.3124843 | 0.3308861 | 0.3497368 | 0.3769788 | 0.3934043 | 0.4193905 | 0.4382476 | 0.4771240 | 0.6033114 | 0.6916735 | 0.7305025 | 0.7337248 |
NDVI | Planted/Cultivated | 97 | 0 | 0.5332255 | 0.1213052 | 0.01231668 | 0.1373269 | 0.5177150 | -0.6469923 | 0.3249635 | 0.3262391 | 0.3740784 | 0.3937608 | 0.4236221 | 0.4498498 | 0.4584951 | 0.4879352 | 0.5132312 | 0.5297822 | 0.5670107 | 0.5871767 | 0.6677204 | 0.7226615 | 0.7493749 | 0.7969922 | 0.7969922 |
NDVI | Shrubland | 130 | 0 | 0.3065798 | 0.1295559 | 0.01136280 | 0.1688918 | 1.1160872 | 0.4843057 | 0.1424335 | 0.1501656 | 0.1663934 | 0.1871671 | 0.2014691 | 0.2101985 | 0.2158819 | 0.2352937 | 0.2691359 | 0.2868186 | 0.3441049 | 0.3790904 | 0.4145865 | 0.5239458 | 0.5461648 | 0.6790463 | 0.6939532 |
SOC | Forest | 93 | 0 | 10.4308817 | 6.8021471 | 0.70534979 | 10.3720000 | 0.7778065 | -0.1552659 | 1.3330000 | 1.3578400 | 2.3656000 | 3.3538000 | 4.4686000 | 4.9310000 | 5.1440000 | 6.5884000 | 8.9740000 | 11.1936000 | 13.7130000 | 15.3030000 | 16.6020000 | 20.8730000 | 22.2096000 | 28.1831200 | 30.4730000 |
SOC | Herbaceous | 150 | 1 | 5.4769667 | 3.9250913 | 0.32048236 | 4.3425000 | 1.2775572 | 1.4077233 | 0.4080000 | 0.5794500 | 1.0895000 | 1.4265000 | 2.2820000 | 2.6240000 | 3.0726000 | 3.5692000 | 4.6090000 | 5.2000000 | 6.3490000 | 6.9665000 | 8.2500000 | 11.0768000 | 13.3133500 | 17.5073200 | 18.8140000 |
SOC | Planted/Cultivated | 97 | 0 | 6.6967216 | 3.5983014 | 0.36535215 | 5.4170000 | 0.5350421 | -0.2587066 | 0.4620000 | 0.7192800 | 1.6380000 | 2.4642000 | 3.6094000 | 4.0020000 | 4.3260000 | 5.3820000 | 6.2300000 | 7.2166000 | 8.0810000 | 9.4190000 | 10.1170000 | 11.3962000 | 13.1846000 | 15.7638400 | 16.3360000 |
SOC | Shrubland | 127 | 3 | 4.1307638 | 3.7448591 | 0.33230251 | 4.3105000 | 1.7061821 | 2.9893653 | 0.4460000 | 0.4746400 | 0.6162000 | 0.8046000 | 1.1344000 | 1.3500000 | 1.6174000 | 2.4386000 | 2.9960000 | 3.7474000 | 4.9050000 | 5.6605000 | 6.2840000 | 9.0786000 | 12.2261000 | 15.8518000 | 19.0990000 |
correlate() calculates the correlation coefficient of all combinations of several numerical variables as follows:
var1 | var2 | coef_corr |
|---|---|---|
DEM | SOC | 0.16668949 |
MAP | SOC | 0.49886194 |
MAT | SOC | -0.35802586 |
NDVI | SOC | 0.58704521 |
SOC | DEM | 0.16668949 |
MAP | DEM | -0.30672789 |
MAT | DEM | -0.80753239 |
NDVI | DEM | -0.06733319 |
SOC | MAP | 0.49886194 |
DEM | MAP | -0.30672789 |
MAT | MAP | 0.06032649 |
NDVI | MAP | 0.80528269 |
SOC | MAT | -0.35802586 |
DEM | MAT | -0.80753239 |
MAP | MAT | 0.06032649 |
NDVI | MAT | -0.20967449 |
SOC | NDVI | 0.58704521 |
DEM | NDVI | -0.06733319 |
MAP | NDVI | 0.80528269 |
MAT | NDVI | -0.20967449 |
The correlate() also supports the group_by() function syntax in the dplyr package.
NLCD | var1 | var2 | coef_corr |
|---|---|---|---|
Forest | DEM | SOC | 0.30298598 |
Forest | MAP | SOC | 0.48134776 |
Forest | MAT | SOC | -0.46258320 |
Forest | NDVI | SOC | 0.39140504 |
Forest | SOC | DEM | 0.30298598 |
Forest | MAP | DEM | 0.41052474 |
Forest | MAT | DEM | -0.71735792 |
Forest | NDVI | DEM | 0.39660074 |
Forest | SOC | MAP | 0.48134776 |
Forest | DEM | MAP | 0.41052474 |
Forest | MAT | MAP | -0.62794270 |
Forest | NDVI | MAP | 0.63598331 |
Forest | SOC | MAT | -0.46258320 |
Forest | DEM | MAT | -0.71735792 |
Forest | MAP | MAT | -0.62794270 |
Forest | NDVI | MAT | -0.48149700 |
Forest | SOC | NDVI | 0.39140504 |
Forest | DEM | NDVI | 0.39660074 |
Forest | MAP | NDVI | 0.63598331 |
Forest | MAT | NDVI | -0.48149700 |
Herbaceous | DEM | SOC | -0.22121181 |
Herbaceous | MAP | SOC | 0.48276882 |
Herbaceous | MAT | SOC | -0.10104174 |
Herbaceous | NDVI | SOC | 0.52751680 |
Herbaceous | SOC | DEM | -0.22121181 |
Herbaceous | MAP | DEM | -0.57025818 |
Herbaceous | MAT | DEM | -0.66962468 |
Herbaceous | NDVI | DEM | -0.59258641 |
Herbaceous | SOC | MAP | 0.48276882 |
Herbaceous | DEM | MAP | -0.57025818 |
Herbaceous | MAT | MAP | 0.35544711 |
Herbaceous | NDVI | MAP | 0.87783385 |
Herbaceous | SOC | MAT | -0.10104174 |
Herbaceous | DEM | MAT | -0.66962468 |
Herbaceous | MAP | MAT | 0.35544711 |
Herbaceous | NDVI | MAT | 0.18936050 |
Herbaceous | SOC | NDVI | 0.52751680 |
Herbaceous | DEM | NDVI | -0.59258641 |
Herbaceous | MAP | NDVI | 0.87783385 |
Herbaceous | MAT | NDVI | 0.18936050 |
Planted/Cultivated | DEM | SOC | -0.31880666 |
Planted/Cultivated | MAP | SOC | 0.42971838 |
Planted/Cultivated | MAT | SOC | 0.06276231 |
Planted/Cultivated | NDVI | SOC | 0.47472055 |
Planted/Cultivated | SOC | DEM | -0.31880666 |
Planted/Cultivated | MAP | DEM | -0.88745536 |
Planted/Cultivated | MAT | DEM | -0.83305530 |
Planted/Cultivated | NDVI | DEM | -0.52616146 |
Planted/Cultivated | SOC | MAP | 0.42971838 |
Planted/Cultivated | DEM | MAP | -0.88745536 |
Planted/Cultivated | MAT | MAP | 0.62195665 |
Planted/Cultivated | NDVI | MAP | 0.71441683 |
Planted/Cultivated | SOC | MAT | 0.06276231 |
Planted/Cultivated | DEM | MAT | -0.83305530 |
Planted/Cultivated | MAP | MAT | 0.62195665 |
Planted/Cultivated | NDVI | MAT | 0.17053661 |
Planted/Cultivated | SOC | NDVI | 0.47472055 |
Planted/Cultivated | DEM | NDVI | -0.52616146 |
Planted/Cultivated | MAP | NDVI | 0.71441683 |
Planted/Cultivated | MAT | NDVI | 0.17053661 |
Shrubland | DEM | SOC | 0.39720120 |
Shrubland | MAP | SOC | 0.44532165 |
Shrubland | MAT | SOC | -0.45936785 |
Shrubland | NDVI | SOC | 0.64602818 |
Shrubland | SOC | DEM | 0.39720120 |
Shrubland | MAP | DEM | 0.29834522 |
Shrubland | MAT | DEM | -0.81913433 |
Shrubland | NDVI | DEM | 0.48180780 |
Shrubland | SOC | MAP | 0.44532165 |
Shrubland | DEM | MAP | 0.29834522 |
Shrubland | MAT | MAP | -0.28844730 |
Shrubland | NDVI | MAP | 0.70639285 |
Shrubland | SOC | MAT | -0.45936785 |
Shrubland | DEM | MAT | -0.81913433 |
Shrubland | MAP | MAT | -0.28844730 |
Shrubland | NDVI | MAT | -0.57829256 |
Shrubland | SOC | NDVI | 0.64602818 |
Shrubland | DEM | NDVI | 0.48180780 |
Shrubland | MAP | NDVI | 0.70639285 |
Shrubland | MAT | NDVI | -0.57829256 |
plot.correlate() visualizes the correlation matrix.
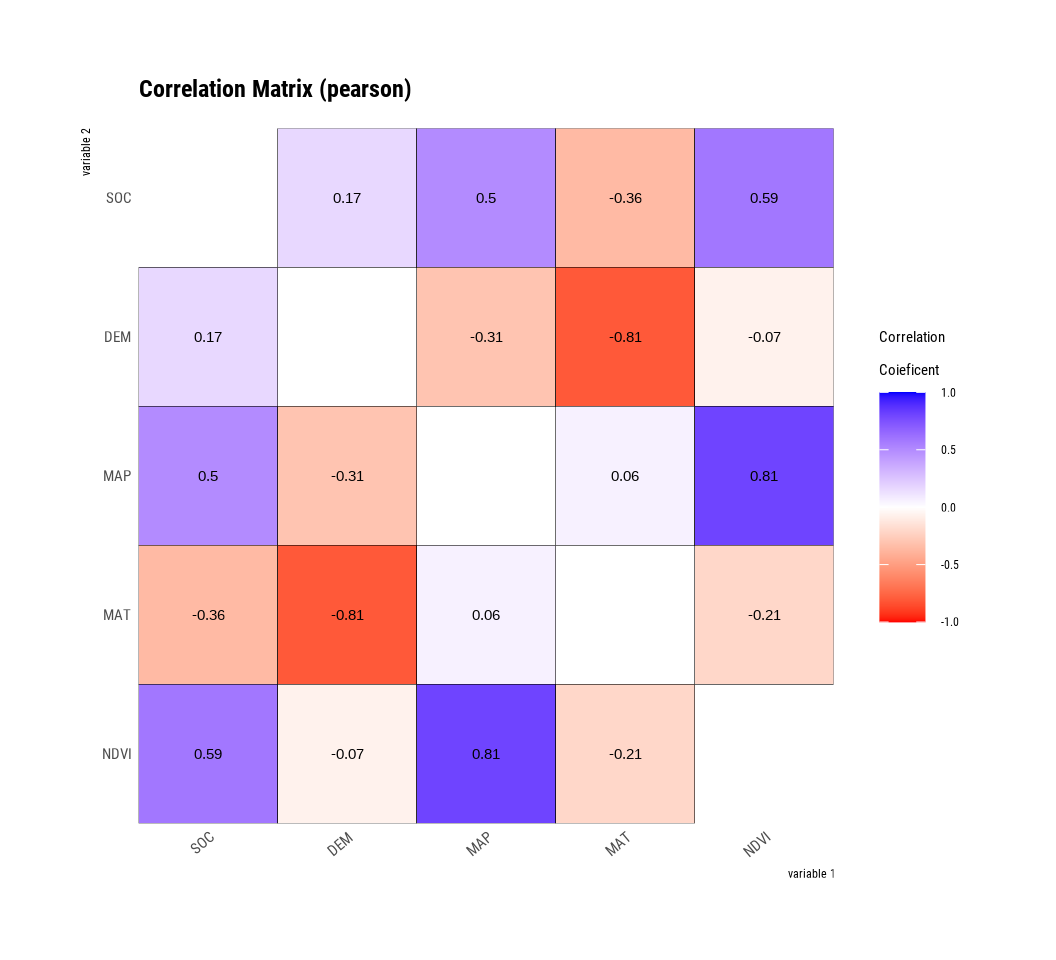
The plot.correlate() function also supports the group_by() function syntax in the dplyr package.
To perform EDA based on the target variable, you must create a target_by class object. target_by() creates a target_by class with an object inheriting data.frame.. target_by() is similar to group_by() in dplyr which creates grouped_df.
The following is an example of specifying VAR as the target variable in as_df data.frame.:
relate() shows the relationship between the target variable and the predictor. The following example shows the relationship between GAs and the target variable VAR. The predictor GAs is a numeric variable. In this case, the descriptive statistics are shown for each level of the target variable.
# A tibble: 8 × 27
described_variables VAR n na mean sd se_mean IQR skewness
<chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 GAs BR01 20 0 1.41 0.485 0.108 0.671 0.387
2 GAs BR06 20 0 1.27 0.484 0.108 0.603 0.619
3 GAs BR28 20 0 1.35 0.516 0.115 0.737 0.278
4 GAs BR35 20 0 1.37 0.454 0.101 0.763 0.194
5 GAs BR36 20 0 1.37 0.526 0.118 0.860 0.476
6 GAs Jefferson 20 0 1.32 0.478 0.107 0.620 0.624
7 GAs Kaybonnet 20 0 1.43 0.546 0.122 0.895 0.0240
8 GAs total 140 0 1.36 0.491 0.0415 0.798 0.345
# ℹ 18 more variables: kurtosis <dbl>, p00 <dbl>, p01 <dbl>, p05 <dbl>,
# p10 <dbl>, p20 <dbl>, p25 <dbl>, p30 <dbl>, p40 <dbl>, p50 <dbl>,
# p60 <dbl>, p70 <dbl>, p75 <dbl>, p80 <dbl>, p90 <dbl>, p95 <dbl>,
# p99 <dbl>, p100 <dbl>plot() visualizes the relate class object created by relate() as the relationship between the target and predictor variables. The relationship between GAs and VAR visualized by a density plot.
The following example shows the relationship between TREAT and the target variable VAR. The predictor variable TREAT is categorical. This case illustrates the contingency table of two variables. The summary() function performs an independence test on the contingency table.
Call: xtabs(formula = formula_str, data = data, addNA = TRUE)
Number of cases in table: 140
Number of factors: 2
Test for independence of all factors:
Chisq = 0, df = 6, p-value = 1plot() visualizes the relationship between the target variable and the predictor. A mosaics plot represents the relationship between VAR and TREAT.
When the numeric variable GY is the target variable, we examine the relationship between the target variable and the predictor.
The following example shows the relationship between GAs and the target variable GY. The predictor variable GAs is numeric. In this case, it shows the result of a simple linear model of the target ~ predictor formula. The ‘summary()’ function expresses the details of the model.
Call:
lm(formula = formula_str, data = data)
Residuals:
Min 1Q Median 3Q Max
-32.308 -5.749 -0.707 6.801 34.242
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 50.983 2.697 18.901 < 2e-16 ***
GAs -16.410 1.866 -8.792 5.23e-15 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 10.8 on 138 degrees of freedom
Multiple R-squared: 0.3591, Adjusted R-squared: 0.3544
F-statistic: 77.31 on 1 and 138 DF, p-value: 5.235e-15plot() visualizes the relationship between the target and predictor variables. The relationship between GY and GAs is visualized with a scatter plot. The figure on the left shows the scatter plot of GY and GAs and the confidence interval of the regression line and regression line. The figure on the right shows the relationship between the original data and the predicted values of the linear model as a scatter plot. If there is a linear relationship between the two variables, the scatter plot of the observations converges on the red diagonal line.
The following example shows the relationship between VAR and the target The predictor VAR is a categorical variable and displays the result of a one-way ANOVA of the target ~ predictor relationship. The results are expressed in terms of ANOVA. The summary() function shows the regression coefficients for each level of the predictor. In other words, it shows detailed information about the simple regression analysis of the target ~ predictor relationship.
dlookr imputes missing values and outliers and resolves skewed data. It also provides the ability to bin continuous variables as categorical variables.
Here is a list of the data conversion functions and functions provided by dlookr:
find_na() finds a variable that contains the missing values variable, and imputate_na() imputes the missing values.
find_outliers() finds a variable that contains the outliers, and imputate_outlier() imputes the outlier.
summary.imputation() and plot.imputation() provide information and visualization of the imputed variables.
find_skewness() finds the variables of the skewed data, and transform() performs the resolving of the skewed data.
transform() also performs standardization of numeric variables.
summary.transform() and plot.transform() provide information and visualization of transformed variables.
binning() and binning_by() convert binational data into categorical data.
print.bins() and summary.bins() show and summarize the binning results.
plot.bins() and plot.optimal_bins() provide visualization of the binning result.
transformation_report() performs the data transform and reports the result.
imputate_na()imputate_na() imputes the missing value contained in the variable. The predictor with missing values supports both numeric and categorical variables and supports the following method.
predictor is numerical variable
“mean”: arithmetic mean
“median”: median
“mode”: mode
“knn”: K-nearest neighbors
“rpart”: Recursive Partitioning and Regression Trees
“mice”: Multivariate Imputation by Chained Equations
target variable must be specified
random seed must be set
predictor is categorical variable
“mode”: mode
“rpart”: Recursive Partitioning and Regression Trees
“mice”: Multivariate Imputation by Chained Equations
target variable must be specified
random seed must be set
In the following example, imputate_na() imputes the missing value of SOC, a numeric variable of mf dataframe, using the “rpart” method. summary() summarizes missing value imputation information, and plot() visualizes missing information.
* Impute missing values based on Recursive Partitioning and Regression Trees
- method : rpart
* Information of Imputation (before vs after)
Original Imputation
described_variables "value" "value"
n "467" "471"
na "4" "0"
mean "6.350762" "6.327249"
sd "5.045409" "5.031445"
se_mean "0.2334737" "0.2318367"
IQR "5.9440" "5.8595"
skewness "1.464728" "1.476371"
kurtosis "2.427192" "2.469712"
p00 "0.408" "0.408"
p01 "0.49094" "0.49130"
p05 "0.9637" "0.9735"
p10 "1.2902" "1.2930"
p20 "2.3294" "2.3350"
p25 "2.7695" "2.7775"
p30 "3.1114" "3.0940"
p40 "3.9906" "3.9740"
p50 "4.971" "4.953"
p60 "6.1266" "6.0980"
p70 "7.503" "7.438"
p75 "8.7135" "8.6370"
p80 "10.0522" " 9.9570"
p90 "13.383" "13.351"
p95 "16.5219" "16.5185"
p99 "22.12476" "22.06820"
p100 "30.473" "30.473" transform()transform() performs data transformation. Only numeric variables are supported, and the following methods are provided.
Standardization
“zscore”: z-score transformation. (x - mu) / sigma
“minmax”: minmax transformation. (x - min) / (max - min)
Resolving Skewness
“log”: log transformation. log(x)
“log+1”: log transformation. log(x + 1). Used for values that contain 0.
“sqrt”: square root transformation.
“1/x”: 1 / x transformation
“x^2”: x square transformation
“x^3”: x^3 square transformation
Use the methods zscore and minmax to perform standardization.
transform()find_skewness() searches for variables with skewed data. This function finds data skewed by search conditions and calculates skewness.
ID FIPS STATE_ID Longitude Latitude SOC DEM Aspect
-0.008 0.326 0.327 0.750 -0.112 1.460 -0.023 0.533
Slope TPI KFactor MAP MAT NDVI SiltClay
1.627 -1.084 -0.540 1.079 -0.274 0.233 0.110 skewness of SOC is 1.46. This means that the distribution of data is inclined to the left. So, for normal distribution, use transform() to convert to the “log” method as follows. summary() summarizes transformation information, and plot() visualizes transformation information.
* Resolving Skewness with log
* Information of Transformation (before vs after)
Original Transformation
n 467.0000000 467.00000000
na 4.0000000 4.00000000
mean 6.3507623 1.51955339
sd 5.0454091 0.87144092
se_mean 0.2334737 0.04032548
IQR 5.9440000 1.14617777
skewness 1.4647284 -0.45555425
kurtosis 2.4271923 -0.16754924
p00 0.4080000 -0.89648810
p01 0.4909400 -0.71147121
p05 0.9637000 -0.03724314
p10 1.2902000 0.25479371
p20 2.3294000 0.84561000
p25 2.7695000 1.01866665
p30 3.1114000 1.13507226
p40 3.9906000 1.38393888
p50 4.9710000 1.60362103
p60 6.1266000 1.81263983
p70 7.5030000 2.01530262
p75 8.7135000 2.16484442
p80 10.0522000 2.30778025
p90 13.3830000 2.59398096
p95 16.5219000 2.80468666
p99 22.1247600 3.09624501
p100 30.4730000 3.41684105binning()binning() transforms a numeric variable into a categorical variable by binning it. The following types of binning are supported.
“quantile”: categorize using quantile to include the same frequencies
“equal”: categorize to have equal length segments
“pretty”: categorized into moderately good segments
“kmeans”: categorization using K-means clustering
“bclust”: categorization using bagged clustering technique
Here are some examples of how to bin SOC using binning().:
binned type: quantile
number of bins: 10
x
[0.408,1.286467] (1.286467,2.3232] (2.3232,3.109267] (3.109267,3.988067]
47 46 47 47
(3.988067,4.971] (4.971,6.1274] (6.1274,7.507] (7.507,10.076]
47 46 47 48
(10.076,13.42567] (13.42567,30.473] <NA>
45 47 4 levels freq rate
1 [0.408,1.286467] 47 0.099787686
2 (1.286467,2.3232] 46 0.097664544
3 (2.3232,3.109267] 47 0.099787686
4 (3.109267,3.988067] 47 0.099787686
5 (3.988067,4.971] 47 0.099787686
6 (4.971,6.1274] 46 0.097664544
7 (6.1274,7.507] 47 0.099787686
8 (7.507,10.076] 48 0.101910828
9 (10.076,13.42567] 45 0.095541401
10 (13.42567,30.473] 47 0.099787686
11 <NA> 4 0.008492569This tutorial explores exploratory data analysis (EDA) using the R-package ‘dlookr.’ You’ll learn about the package’s features, such as missing data analysis and outlier detection, and functions that facilitate the generation of insightful reports and visualizations. Additionally, you’ll learn how ‘dlookr’ contributes to a comprehensive understanding of data quality. Remember to consider exploring its additional functionalities, practice with diverse datasets, and adapt the techniques to the unique characteristics of your data.