Code
library(tidyverse)
library(tidymodels)
library(Metrics)
library(DT)
In order for machine learning software to truly be accessible to non-experts, H2O have designed an easy-to-use interface which automates the process of training a large selection of candidate models. H2O’s AutoML can also be a helpful tool for the advanced user, by providing a simple wrapper function that performs a large number of modeling-related tasks that would typically require many lines of code, and by freeing up their time to focus on other aspects of the data science pipeline tasks such as data-preprocessing, feature engineering and model deployment.
H2O’s AutoML can be used for automating the machine learning workflow, which includes automatic training and tuning of many models within a user-specified time-limit.
H2O offers a number of model explainability methods that apply to AutoML objects (groups of models), as well as individual models (e.g. leader model). Explanations can be generated automatically with a single function call, providing a simple interface to exploring and explaining the AutoML models.
In this exercise we will use following synthetic data set and use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
Rows: 1,408
Columns: 9
$ SOC <dbl> 1.900, 2.644, 0.800, 0.736, 15.641, 8.818, 3.782, 6.641, 4.803, …
$ DEM <dbl> 2825.1111, 2535.1086, 1716.3300, 1649.8933, 2675.3113, 2581.4839…
$ Slope <dbl> 18.981682, 14.182393, 1.585145, 9.399726, 12.569353, 6.358553, 1…
$ TPI <dbl> -0.91606224, -0.15259802, -0.39078590, -2.54008722, 7.40076303, …
$ MAT <dbl> 4.709227, 4.648000, 6.360833, 10.265385, 2.798550, 6.358550, 7.0…
$ MAP <dbl> 613.6979, 597.7912, 201.5091, 298.2608, 827.4680, 679.1392, 508.…
$ NDVI <dbl> 0.6845260, 0.7557631, 0.2215059, 0.2785148, 0.7337426, 0.7017139…
$ NLCD <chr> "Forest", "Forest", "Shrubland", "Shrubland", "Forest", "Forest"…
$ FRG <chr> "Fire Regime Group IV", "Fire Regime Group IV", "Fire Regime Gro…
H2O is not running yet, starting it now...
Note: In case of errors look at the following log files:
C:\Users\zahmed2\AppData\Local\Temp\1\RtmpAZaiK7\file7f1459af2a3f/h2o_zahmed2_started_from_r.out
C:\Users\zahmed2\AppData\Local\Temp\1\RtmpAZaiK7\file7f147da41165/h2o_zahmed2_started_from_r.err
Starting H2O JVM and connecting: Connection successful!
R is connected to the H2O cluster:
H2O cluster uptime: 2 seconds 945 milliseconds
H2O cluster timezone: America/New_York
H2O data parsing timezone: UTC
H2O cluster version: 3.40.0.4
H2O cluster version age: 3 months and 23 days
H2O cluster name: H2O_started_from_R_zahmed2_yqz807
H2O cluster total nodes: 1
H2O cluster total memory: 148.00 GB
H2O cluster total cores: 40
H2O cluster allowed cores: 40
H2O cluster healthy: TRUE
H2O Connection ip: localhost
H2O Connection port: 54321
H2O Connection proxy: NA
H2O Internal Security: FALSE
R Version: R version 4.3.1 (2023-06-16 ucrt) The H2O AutoML interface is designed to have as few parameters as possible so that all the user needs to do is point to their dataset, identify the response column and optionally specify a time constraint or limit on the number of total models trained.
In both the R and Python API, AutoML uses the same data-related arguments, x, y, training_frame, validation_frame, as the other H2O algorithms. Most of the time, all you’ll need to do is specify the data arguments. You can then configure values for max_runtime_secs and/or max_models to set explicit time or number-of-model limits on your run.
y: This argument is the name (or index) of the response column.
training_frame: Specifies the training set.
One of the following stopping strategies (time or number-of-model based) must be specified. When both options are set, then the AutoML run will stop as soon as it hits one of either When both options are set, then the AutoML run will stop as soon as it hits either of these limits.
max_runtime_secs: This argument specifies the maximum time that the AutoML process will run for. The default is 0 (no limit), but dynamically sets to 1 hour if none of max_runtime_secs and max_models are specified by the user.
max_models: Specify the maximum number of models to build in an AutoML run, excluding the Stacked Ensemble models. Defaults to NULL/None. Always set this parameter to ensure AutoML reproducibility: all models are then trained until convergence and none is constrained by a time budget.
22:23:49.973: Stopping tolerance set by the user is < 70% of the recommended default of 0.026650089544451302, so models may take a long time to converge or may not converge at all.
22:23:49.987: AutoML: XGBoost is not available; skipping it.AutoML Details
==============
Project Name: AutoML_trainings
Leader Model ID: StackedEnsemble_AllModels_1_AutoML_1_20230820_222349
Algorithm: stackedensemble
Total Number of Models Trained: 102
Start Time: 2023-08-20 22:23:50 UTC
End Time: 2023-08-20 22:25:36 UTC
Duration: 106 s
Leaderboard
===========
model_id rmse mse
1 StackedEnsemble_AllModels_1_AutoML_1_20230820_222349 1.602884 2.569237
2 GBM_3_AutoML_1_20230820_222349 1.619452 2.622626
3 StackedEnsemble_BestOfFamily_1_AutoML_1_20230820_222349 1.624302 2.638357
4 GBM_5_AutoML_1_20230820_222349 1.627121 2.647524
5 GBM_grid_1_AutoML_1_20230820_222349_model_17 1.653648 2.734552
6 GBM_grid_1_AutoML_1_20230820_222349_model_34 1.655886 2.741959
7 GBM_grid_1_AutoML_1_20230820_222349_model_3 1.666404 2.776901
8 GBM_4_AutoML_1_20230820_222349 1.672820 2.798326
9 GBM_grid_1_AutoML_1_20230820_222349_model_31 1.725521 2.977424
10 GBM_grid_1_AutoML_1_20230820_222349_model_5 1.735791 3.012971
mae rmsle mean_residual_deviance
1 0.7222105 0.2536849 2.569237
2 0.7239151 0.2622542 2.622626
3 0.7476178 0.2620803 2.638357
4 0.7039202 0.2562144 2.647524
5 0.7593434 0.2707456 2.734552
6 0.6850420 0.2643083 2.741959
7 0.8903464 0.2730299 2.776901
8 0.7283962 0.2664887 2.798326
9 0.8773841 0.2799215 2.977424
10 0.8666020 0.2818231 3.012971
[102 rows x 6 columns] Model Details:
==============
H2ORegressionModel: stackedensemble
Model ID: StackedEnsemble_AllModels_1_AutoML_1_20230820_222349
Model Summary for Stacked Ensemble:
key value
1 Stacking strategy cross_validation
2 Number of base models (used / total) 9/100
3 # GBM base models (used / total) 9/42
4 # DRF base models (used / total) 0/2
5 # DeepLearning base models (used / total) 0/55
6 # GLM base models (used / total) 0/1
7 Metalearner algorithm GLM
8 Metalearner fold assignment scheme Random
9 Metalearner nfolds 5
10 Metalearner fold_column NA
11 Custom metalearner hyperparameters None
H2ORegressionMetrics: stackedensemble
** Reported on training data. **
MSE: 0.04266853
RMSE: 0.2065636
MAE: 0.1490082
RMSLE: 0.04660908
Mean Residual Deviance : 0.04266853
H2ORegressionMetrics: stackedensemble
** Reported on cross-validation data. **
** 5-fold cross-validation on training data (Metrics computed for combined holdout predictions) **
MSE: 2.569237
RMSE: 1.602884
MAE: 0.7222105
RMSLE: 0.2536849
Mean Residual Deviance : 2.569237
Cross-Validation Metrics Summary:
mean sd cv_1_valid cv_2_valid
mae 0.719265 0.124818 0.912796 0.671731
mean_residual_deviance 2.555708 1.322591 4.771241 1.623622
mse 2.555708 1.322591 4.771241 1.623622
null_deviance 7123.776400 1225.410400 8354.074000 6459.341000
r2 0.899019 0.043828 0.838288 0.930638
residual_deviance 719.766700 373.211730 1345.490000 446.496150
rmse 1.561806 0.381561 2.184317 1.274214
rmsle 0.253178 0.027742 0.275726 0.252383
cv_3_valid cv_4_valid cv_5_valid
mae 0.590403 0.764795 0.656603
mean_residual_deviance 1.701732 2.787133 1.894810
mse 1.701732 2.787133 1.894810
null_deviance 6337.831500 5928.159700 8539.476000
r2 0.921250 0.867262 0.937656
residual_deviance 496.905760 783.184300 526.757260
rmse 1.304504 1.669471 1.376521
rmsle 0.224472 0.285990 0.227318[1] "RMSE: 4.35"[1] "MAE: 3"library(ggpmisc)
formula<-y~x
ggplot(test.xy, aes(SOC,Pred_SOC)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("Best-ML") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))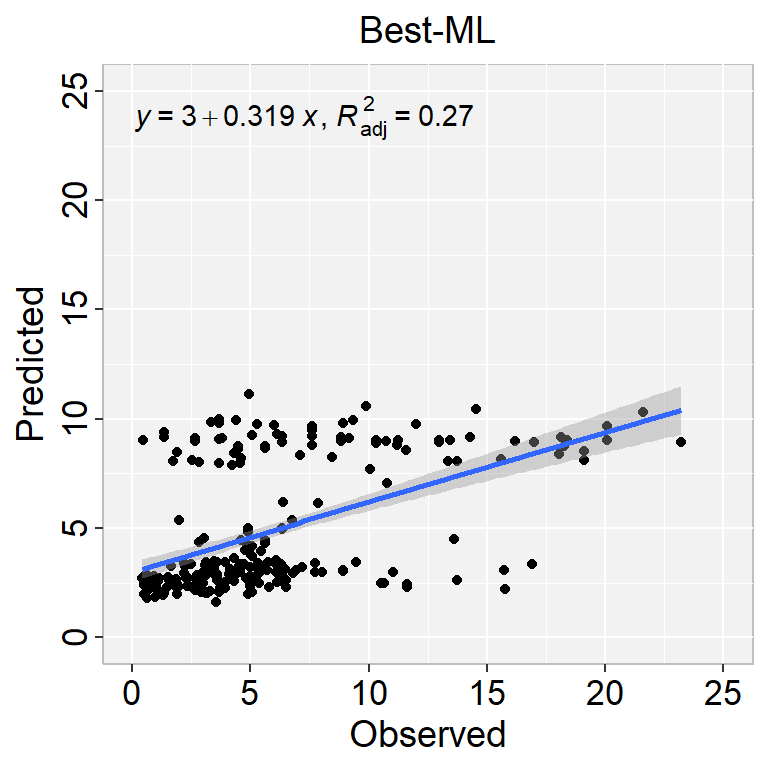
[{fig-align="left" width="254"}](https://github.com/zia207/r-colab/blob/main/autoML_h2o.ipynb)
# Automatic Machine Learning (AutoML) with H20 {.unnumbered}

In order for machine learning software to truly be accessible to non-experts, H2O have designed an easy-to-use interface which automates the process of training a large selection of candidate models. H2O's AutoML can also be a helpful tool for the advanced user, by providing a simple wrapper function that performs a large number of modeling-related tasks that would typically require many lines of code, and by freeing up their time to focus on other aspects of the data science pipeline tasks such as data-preprocessing, feature engineering and model deployment.
H2O's AutoML can be used for automating the machine learning workflow, which includes automatic training and tuning of many models within a user-specified time-limit.
H2O offers a number of model explainability methods that apply to AutoML objects (groups of models), as well as individual models (e.g. leader model). Explanations can be generated automatically with a single function call, providing a simple interface to exploring and explaining the AutoML models.
#### Load Library
```{r}
#| warning: false
#| error: false
library(tidyverse)
library(tidymodels)
library(Metrics)
library(DT)
```
#### Data
In this exercise we will use following synthetic data set and use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
[gp_soil_data_syn.csv](https://www.dropbox.com/s/c63etg7u5qql2y8/gp_soil_data_syn.csv?dl=0)
```{r}
#| warning: false
#| error: false
#| fig.width: 5
#| fig.height: 4
# define file from my github
urlfile = "https://github.com//zia207/r-colab/raw/main/Data/USA/gp_soil_data_syn.csv"
mf<-read_csv(url(urlfile))
# Create a data-frame
df<-mf %>% dplyr::select(SOC, DEM, Slope, TPI,MAT, MAP,NDVI, NLCD, FRG)%>%
glimpse()
```
#### Convert to factor
```{r}
#| warning: false
#| error: false
df$NLCD <- as.factor(df$NLCD)
df$FRG <- as.factor(df$FRG)
```
#### Data split
```{r}
#| warning: false
#| error: false
set.seed(1245) # for reproducibility
split_01 <- initial_split(df, prop = 0.8, strata = SOC)
train <- split_01 %>% training()
test <- split_01 %>% testing()
```
#### Data Scaling
```{r}
train[-c(1, 8,9)] = scale(train[-c(1,8,9)])
test[-c(1, 8,9)] = scale(test[-c(1,8,9)])
```
#### Import h2o
```{r}
#| warning: false
#| error: false
library(h2o)
h2o.init(nthreads = -1, max_mem_size = "148g", enable_assertions = FALSE)
#disable progress bar for RMarkdown
h2o.no_progress()
# Optional: remove anything from previous session
h2o.removeAll()
```
#### Import data to h2o cluster
```{r}
#| warning: false
#| error: false
h_df=as.h2o(df)
h_train = as.h2o(train)
h_test = as.h2o(test)
```
```{r}
train.xy<- as.data.frame(h_train)
test.xy<- as.data.frame(h_test)
```
#### Define response and predictors
```{r}
#| warning: false
#| error: false
y <- "SOC"
x <- setdiff(names(h_df), y)
```
#### AutoML: Automatic Machine Learning
The H2O AutoML interface is designed to have as few parameters as possible so that all the user needs to do is point to their dataset, identify the response column and optionally specify a time constraint or limit on the number of total models trained.
In both the R and Python API, AutoML uses the same data-related arguments, `x`, `y`, `training_frame`, `validation_frame`, as the other H2O algorithms. Most of the time, all you'll need to do is specify the data arguments. You can then configure values for `max_runtime_secs` and/or `max_models` to set explicit time or number-of-model limits on your run.
#### **Required Data Parameters**
- [y](https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/algo-params/y.html): This argument is the name (or index) of the response column.
- [training_frame](https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/algo-params/training_frame.html): Specifies the training set.
#### **Required Stopping Parameters**
One of the following stopping strategies (time or number-of-model based) must be specified. When both options are set, then the AutoML run will stop as soon as it hits one of either When both options are set, then the AutoML run will stop as soon as it hits either of these limits.
- [max_runtime_secs](https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/algo-params/max_runtime_secs.html): This argument specifies the maximum time that the AutoML process will run for. The default is 0 (no limit), but dynamically sets to 1 hour if none of `max_runtime_secs` and `max_models` are specified by the user.
- [max_models](https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/algo-params/max_models.html): Specify the maximum number of models to build in an AutoML run, excluding the Stacked Ensemble models. Defaults to `NULL/None`. Always set this parameter to ensure AutoML reproducibility: all models are then trained until convergence and none is constrained by a time budget.
```{r}
#| warning: false
#| error: false
df.aml <- h2o.automl(
x= x,
y = y,
training_frame = h_df,
nfolds=5,
keep_cross_validation_predictions = TRUE,
stopping_metric = "RMSE",
stopping_tolerance = 0.001,
stopping_rounds = 3,
max_runtime_secs_per_model = 1,
seed = 42,
max_models = 100,
project_name = "AutoML_trainings")
df.aml
```
### Leaderboard
```{r message=F, warning=F,results = "hide"}
lb <- h2o.get_leaderboard(object = df.aml, extra_columns = "ALL")
datatable(as.data.frame(lb),
rownames = FALSE, options = list(pageLength = 10, scrollX = TRUE, round)) %>%
formatRound(columns = -1, digits = 4)
#write.csv(as.data.frame(lb), "Model_Stat_AutoML.csv", row.names = F)
```
### The best Model
```{r}
best_ML<- h2o.get_best_model(df.aml, criterion = "RMSE" )
best_ML
```
### Prediction
```{r}
best.test<-as.data.frame(h2o.predict(object = best_ML, newdata = h_test))
test.xy$Pred_SOC<-best.test$predict
```
```{r}
RMSE<- Metrics::rmse(test.xy$SOC, test.xy$Pred_SOC)
MAE<- Metrics::mae(test.xy$SOC, test.xy$Pred_SOC)
# Print results
paste0("RMSE: ", round(RMSE,2))
paste0("MAE: ", round(MAE,2))
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 4
library(ggpmisc)
formula<-y~x
ggplot(test.xy, aes(SOC,Pred_SOC)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("Best-ML") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))
```