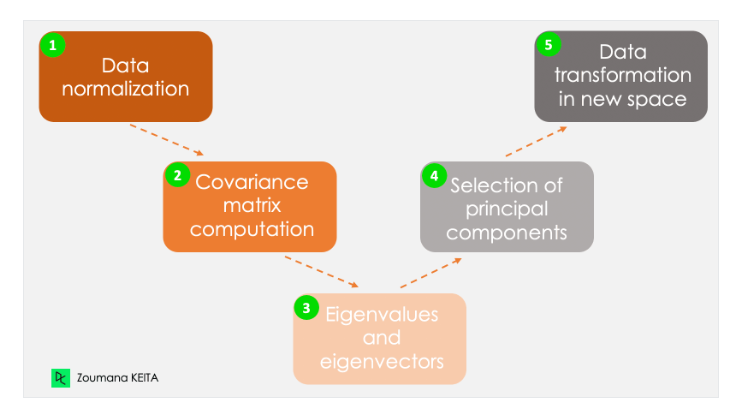
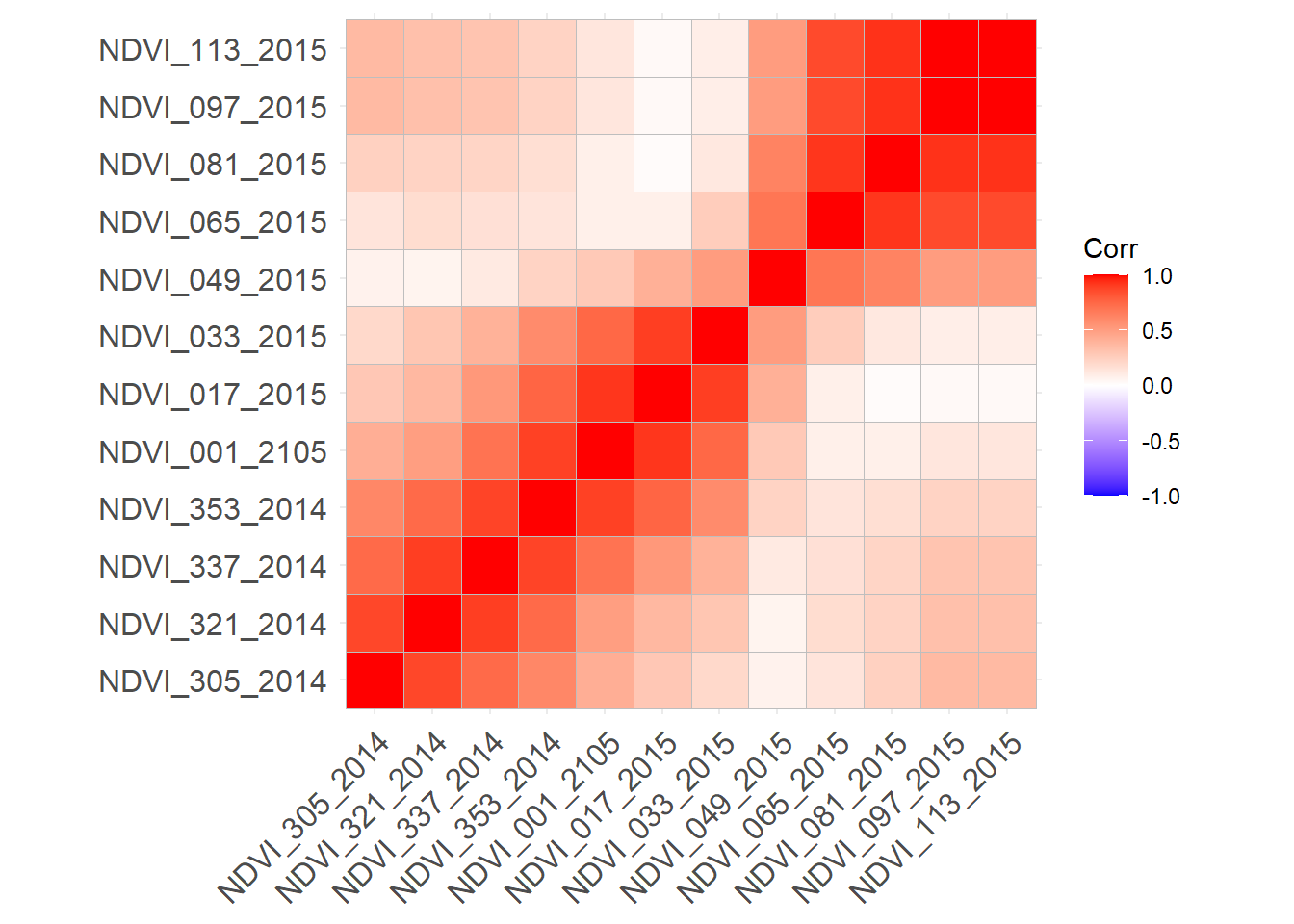
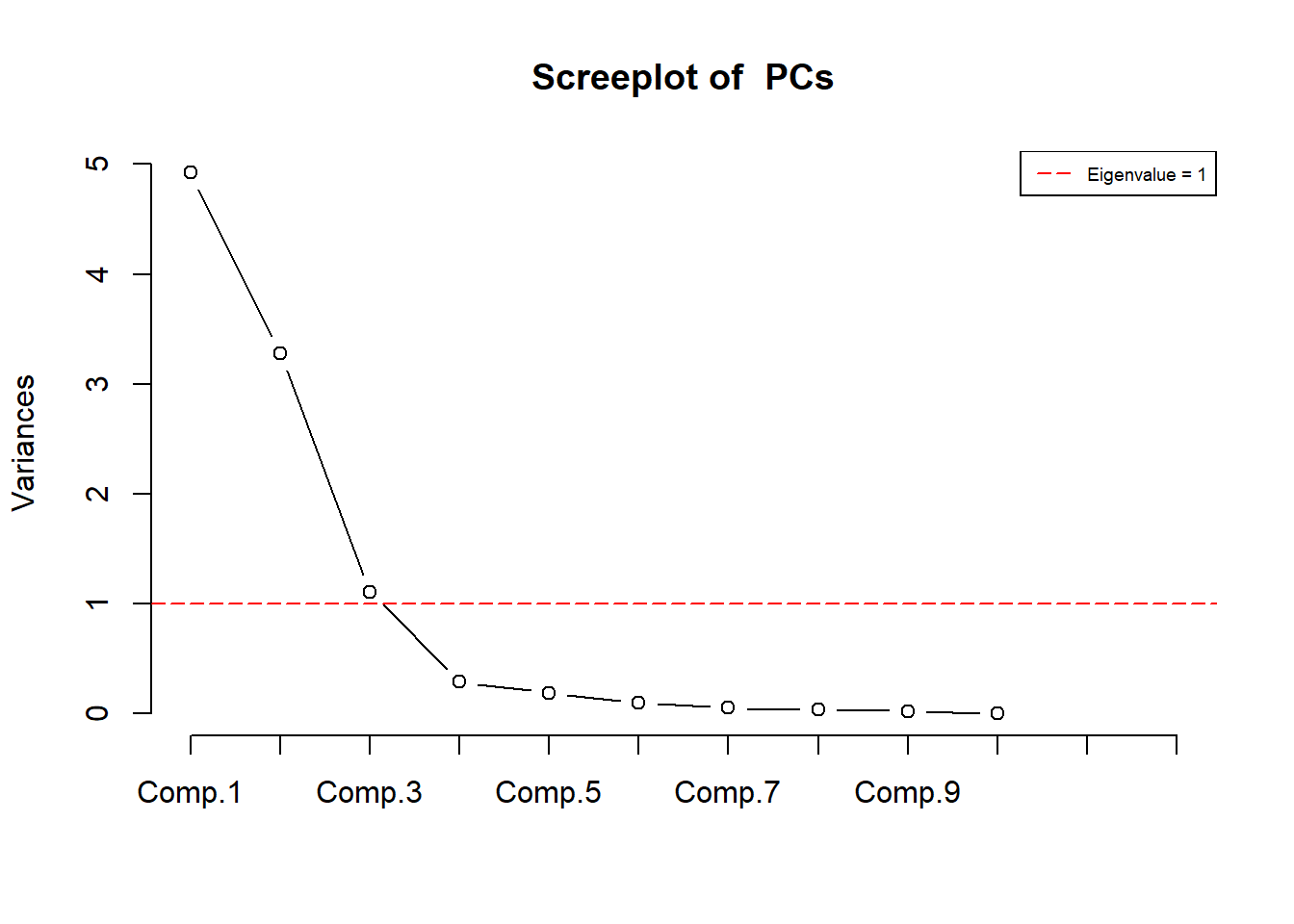
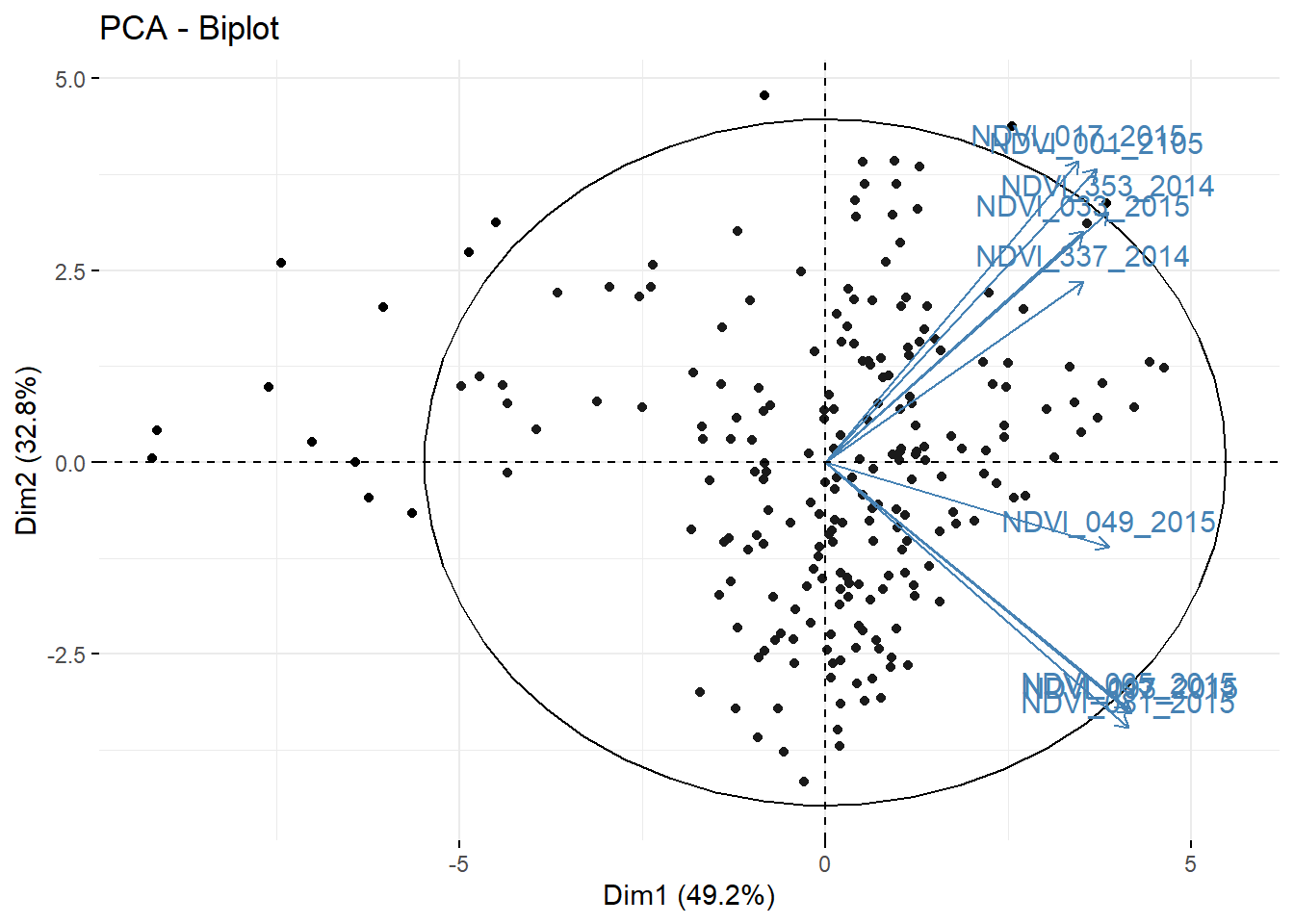
Rows: 223
Columns: 21
$ UNION_ID <dbl> 10040907, 10061031, 10063613, 10065177, 10069494, 100918…
$ x <dbl> 519852.2, 513008.6, 556755.4, 541836.9, 516049.7, 568765…
$ y <dbl> 2439890, 2524428, 2538580, 2508902, 2527786, 2511181, 24…
$ LONG <dbl> 90.1924, 90.1268, 90.5536, 90.4073, 90.1564, 90.6696, 90…
$ LAT <dbl> 22.0637, 22.8275, 22.9544, 22.6868, 22.8578, 22.7065, 22…
$ DIVISION <chr> "Barisal", "Barisal", "Barisal", "Barisal", "Barisal", "…
$ DISTRICT <chr> "Barguna", "Barisal", "Barisal", "Barisal", "Barisal", "…
$ UPAZILA <chr> "Amtali", "Banaripara", "Hizla", "Barisal", "Wazirpur", …
$ UNION <chr> "Amtali", "Bisarkandi", "Bara Jalia", "Roy Pasha Karapur…
$ NDVI_305_2014 <dbl> 0.769, 0.693, 0.675, 0.782, 0.727, 0.593, 0.772, 0.765, …
$ NDVI_321_2014 <dbl> 0.601, 0.572, 0.552, 0.653, 0.587, 0.478, 0.569, 0.590, …
$ NDVI_337_2014 <dbl> 0.549, 0.553, 0.520, 0.673, 0.557, 0.446, 0.543, 0.518, …
$ NDVI_353_2014 <dbl> 0.448, 0.612, 0.518, 0.639, 0.563, 0.405, 0.492, 0.396, …
$ NDVI_001_2105 <dbl> 0.426, 0.615, 0.542, 0.605, 0.554, 0.418, 0.485, 0.364, …
$ NDVI_017_2015 <dbl> 0.414, 0.606, 0.506, 0.579, 0.536, 0.423, 0.494, 0.345, …
$ NDVI_033_2015 <dbl> 0.409, 0.608, 0.508, 0.561, 0.560, 0.452, 0.508, 0.326, …
$ NDVI_049_2015 <dbl> 0.418, 0.640, 0.539, 0.632, 0.660, 0.496, 0.508, 0.415, …
$ NDVI_065_2015 <dbl> 0.424, 0.690, 0.570, 0.671, 0.708, 0.528, 0.497, 0.481, …
$ NDVI_081_2015 <dbl> 0.469, 0.696, 0.563, 0.724, 0.741, 0.518, 0.509, 0.582, …
$ NDVI_097_2015 <dbl> 0.504, 0.677, 0.603, 0.726, 0.714, 0.532, 0.555, 0.648, …
$ NDVI_113_2015 <dbl> 0.504, 0.677, 0.603, 0.726, 0.714, 0.532, 0.555, 0.648, …