Code
library (terra)
library(sf)
library (rgdal)
library(tidyverse)
library(rgeos)
library(maptools)
library(classInt)
library(RColorBrewer)
library(raster) Raster data processing in R is commonly done using packages such as raster and rgdal. Raster data consists of grids of cells, where each cell holds a value representing some information, such as elevation, temperature, or land cover type. The raster package provides functions for reading, manipulating, and analyzing raster data, while the rgdal package focuses on reading and writing geospatial raster data formats.
The terra package is an advanced geospatial package in R that provides powerful tools for working with raster data. It is designed to handle large datasets efficiently and offers a wide range of capabilities for raster data processing, analysis, and visualization. terra is considered a successor to the raster package and aims to provide better performance and support for more complex operations.
Here are some key features and concepts of the terra package:
Efficiency and Performance: terra is optimized for performance, making it suitable for processing large raster datasets. It leverages modern computing hardware and parallel processing capabilities to efficiently handle computations.
Raster Objects: terra introduces the concept of “SpatioTemporalRaster” objects, which are used to represent raster data along with its spatial and temporal attributes.
Basic Operations: Similar to the raster package, terra allows you to perform basic operations like reading, writing, displaying, and extracting values from raster data.
Spatial and Temporal Operations: terra supports spatial and temporal operations such as cropping, reprojecting, resampling, masking, and aggregating raster data. It also handles multi-layer raster stacks efficiently.
Advanced Analytics: The package provides functions for more advanced spatial analytics, including focal operations (neighborhood analysis), zonal statistics, and mathematical operations on raster layers.
Memory Management: terra uses a “lazy” evaluation approach, which means that computations are performed only when needed. This allows for more memory-efficient processing of large datasets.
Visualization: The package includes visualization functions to help you create maps and plots of raster data.
Integration with Other Packages: terra can be integrated with other geospatial packages like sf for vector data handling and analysis.
In this exerciser we will learn following raster processing techniques with R:
Basic Raster Operation
Clipping
Reclassification
Focal Statistics
Raster Algebra
Aggregation
Disaggregation
Resample
Mosaic
Vector to raster conversion
Convert Raster to Point Data
Convert Point Data to Raster
Raster Stack and Raster Brick
We will use SRTM elevation data (250 m x 250 m) of all hilly districts of Bangladesh and The data can be found here for download.
We will use rast() function of terra package to load raster in R
We can load data from github:
You can also load data with raster packages, but raster object relatively slow to perform some tasks. The terra library is more recent (but faster). We can convert a RasterLayer (i.e. a raster object) into a SpatRaster (i.e. a terra object):
class : RasterLayer
dimensions : 1329, 521, 692409 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 648050.4, 778300.4, 2295017, 2627267 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=0 +datum=WGS84 +units=m +no_defs
source : hilly_DEM_UTM.tif
names : Band_1
values : -7.796429, 985.0496 (min, max)class : SpatRaster
dimensions : 1329, 521, 1 (nrow, ncol, nlyr)
resolution : 250, 250 (x, y)
extent : 648050.4, 778300.4, 2295017, 2627267 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=0 +datum=WGS84 +units=m +no_defs
source : hilly_DEM_UTM.tif
name : Band_1
min value : -7.796429
max value : 985.049622 [1] "PROJCRS[\"WGS_1984_Transverse_Mercator\","
[2] " BASEGEOGCRS[\"WGS 84\","
[3] " DATUM[\"World Geodetic System 1984\","
[4] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[5] " LENGTHUNIT[\"metre\",1]]],"
[6] " PRIMEM[\"Greenwich\",0,"
[7] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[8] " ID[\"EPSG\",4326]],"
[9] " CONVERSION[\"Transverse Mercator\","
[10] " METHOD[\"Transverse Mercator\","
[11] " ID[\"EPSG\",9807]],"
[12] " PARAMETER[\"Latitude of natural origin\",0,"
[13] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[14] " ID[\"EPSG\",8801]],"
[15] " PARAMETER[\"Longitude of natural origin\",90,"
[16] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[17] " ID[\"EPSG\",8802]],"
[18] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[19] " SCALEUNIT[\"unity\",1],"
[20] " ID[\"EPSG\",8805]],"
[21] " PARAMETER[\"False easting\",500000,"
[22] " LENGTHUNIT[\"metre\",1],"
[23] " ID[\"EPSG\",8806]],"
[24] " PARAMETER[\"False northing\",0,"
[25] " LENGTHUNIT[\"metre\",1],"
[26] " ID[\"EPSG\",8807]]],"
[27] " CS[Cartesian,2],"
[28] " AXIS[\"easting\",east,"
[29] " ORDER[1],"
[30] " LENGTHUNIT[\"metre\",1,"
[31] " ID[\"EPSG\",9001]]],"
[32] " AXIS[\"northing\",north,"
[33] " ORDER[2],"
[34] " LENGTHUNIT[\"metre\",1,"
[35] " ID[\"EPSG\",9001]]]]" class : SpatRaster
dimensions : 1329, 521, 1 (nrow, ncol, nlyr)
resolution : 250, 250 (x, y)
extent : 648050.4, 778300.4, 2295017, 2627267 (xmin, xmax, ymin, ymax)
coord. ref. : WGS_1984_Transverse_Mercator
source : hilly_DEM_UTM.tif
name : Band_1
min value : -7.796429
max value : 985.049622 R allow you to calculate some basic statistics of raster data. We can use summary function to get some statistics of raster layer.
R-base function hist() is also useful to know frequency of raster values
We can plot raster in diffrent ways in R. R base function plot() can easily apply on raster object for visualization:
R has an image() function that allows you to control the way a raster is rendered on the screen. The plot() function in R has a base setting for the number of pixels that it will plot (100,000 pixels). The image command thus might be better for rendering larger rasters.
To visualize raster in R using ggplot2, first we need to convert it to a dataframe. The terra package has an built-in function for conversion to a plotable dataframe. Then we can applly geom_raster() of ggplot().
Rows: 310,665
Columns: 3
$ x <dbl> 720925.4, 720175.4, 720425.4, 720675.4, 720925.4, 721175.4, 719…
$ y <dbl> 2627142, 2626892, 2626892, 2626892, 2626892, 2626892, 2626642, …
$ Band_1 <dbl> 357.0371, 318.1637, 320.6357, 317.4960, 355.9710, 440.9489, 251…Raster clipping refers to the process of extracting a portion of a raster dataset based on a specified spatial extent or boundary. This is a common operation in geospatial analysis when you need to focus on a specific area of interest within a larger raster dataset. Raster clipping can help reduce the dataset’s size, improve processing efficiency, and facilitate analysis that’s relevant to a particular region.
In R, you can perform raster clipping using various packages, including raster, terra, and others. I’ll provide examples using both the raster and terra packages.
In this exercise, we will clip hilly area DEM with extent of Bandarban district shape file. First we have to create a shape file of this district:
Reading layer `bd_district_BTM' from data source
`/vsicurl/https://github.com//zia207/r-colab/raw/main/Data/Bangladesh//Shapefiles//bd_district_BTM.shp'
using driver `ESRI Shapefile'
Simple feature collection with 64 features and 14 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: 298487.8 ymin: 278578.1 xmax: 778101.8 ymax: 946939.2
Projected CRS: +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defsBefore clipping, you have to make sure both raster and vector data are in same projection systems.
It looks like DEM in UTM and district shape files is BUTM project system. We have bring them in same projection system before clipping. Here we reproject DEM to BUTM using district shape files
In R, clipping of a raster is two steps procedure, first you have to apply crop() function & then mask functions of raster package. crop returns a geographic subset of an object as specified by an extent object (or object from which an extent object can be extracted/created). mask Create a new Raster object that has the same values as input raster.

We can write raster using terra::writeRaster function:
Raster reclassification is the process of assigning new values to the cells of a raster dataset based on predefined criteria. This operation is commonly used in geospatial analysis to reclassify continuous raster data (e.g., elevation, temperature) into discrete categories (e.g., low, medium, high).
In R, you can perform raster reclassification using packages like raster or terra packages
Before reclassification, we will create a histogram with 5 breaks, of dem raster
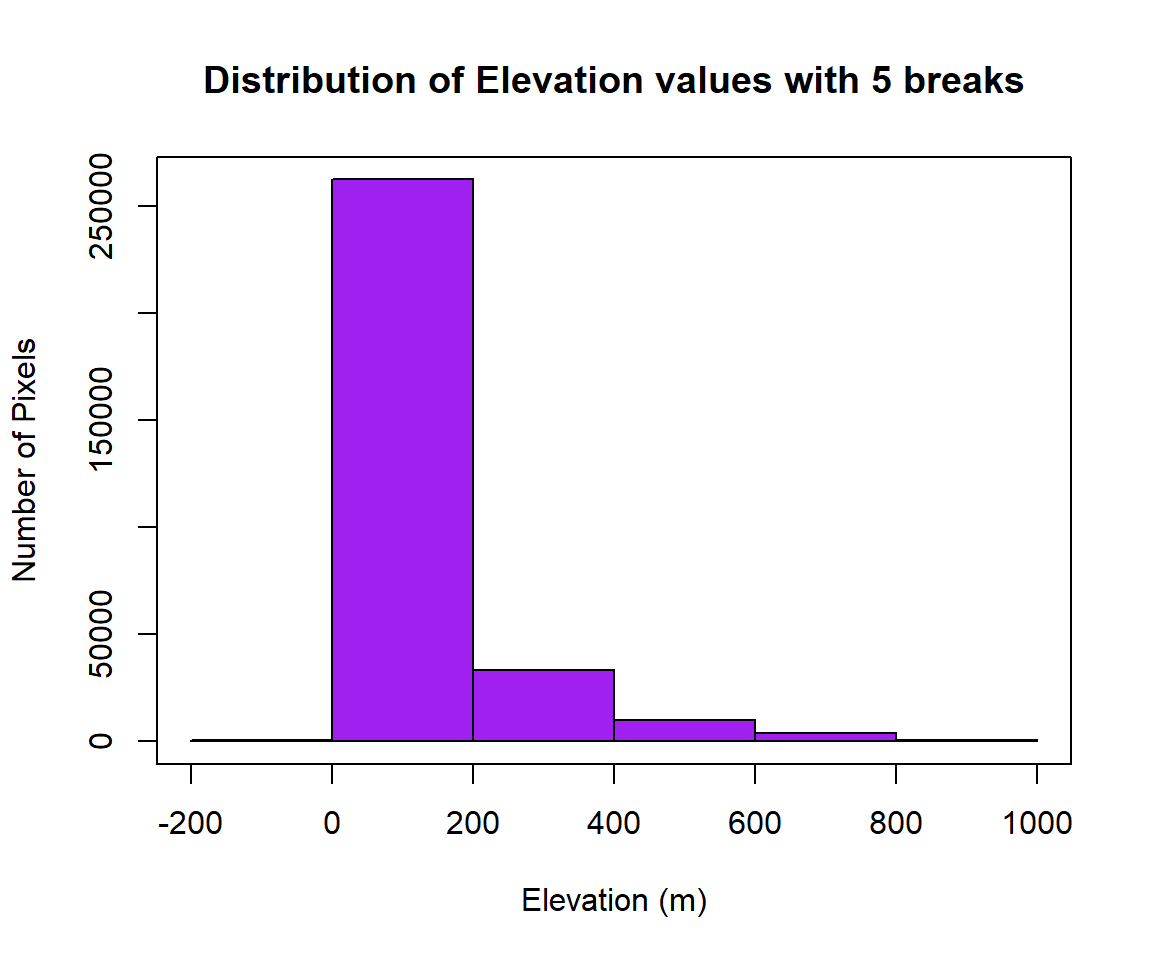
Now, you can check where are breaks and how many pixels in each category, and use these breaks to plot a raster layer
[1] -200 0 200 400 600 800 1000[1] 778 262373 33316 10008 3775 415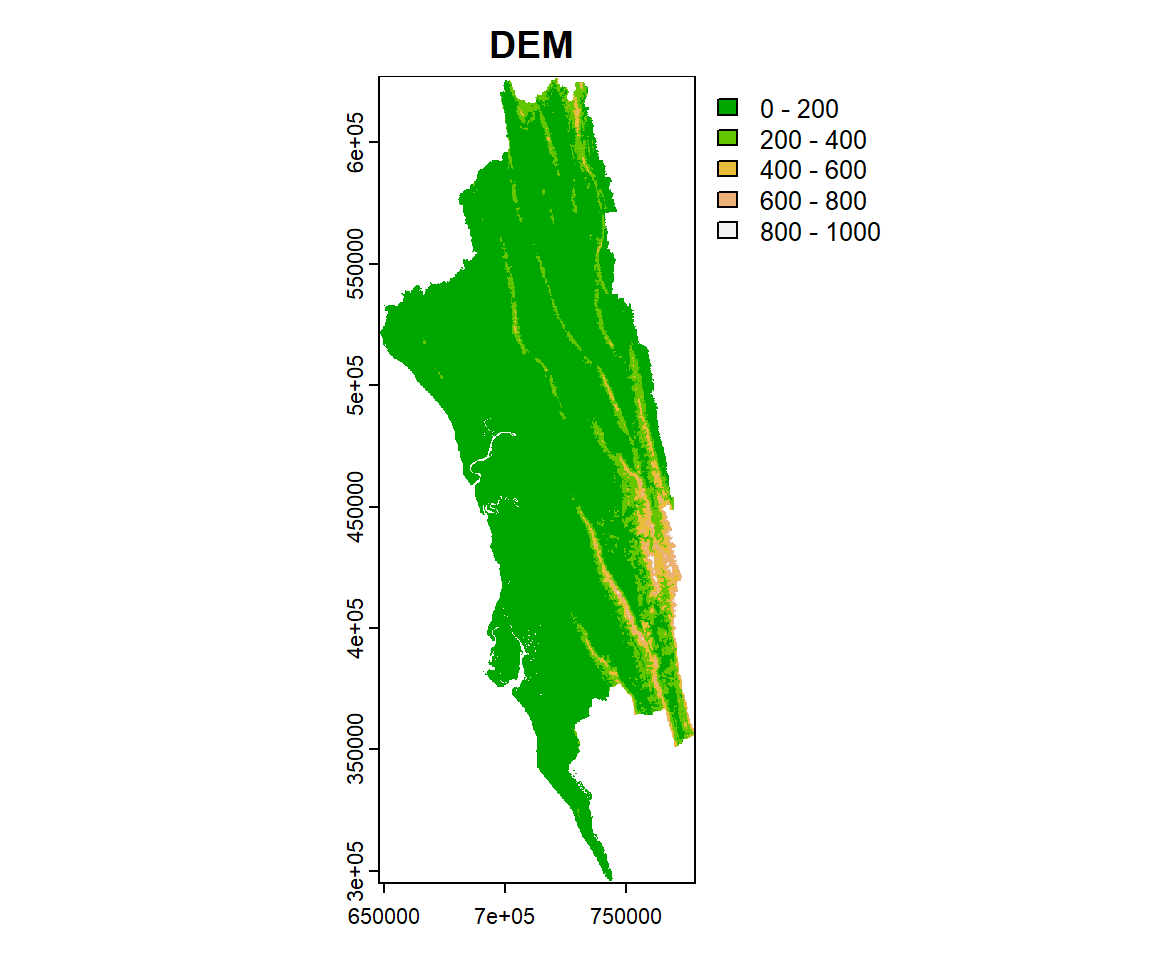
We will use following breaks of histogram to reclassify the dem raster. First, you have to create classification matrix
[1] -200 200 1 200 400 2 400 Inf 3Next, you have to reshape the data frame to matrix using matrix function
[,1] [,2] [,3]
[1,] -200 200 1
[2,] 200 400 2
[3,] 400 Inf 3Now you have to apply terra::classify() function of raster package using re-class_m object.
You can view the distribution of pixels assigned to each class using the bar-plot().
Now we plot the reclassified raster using plot function with a nice looking legend
If you want create a raster map using the quantile, you fave to use classInt package. The classIntervals function is create a custom breaks of raster values using different styles (such as “fixed”, “sd”, “equal”, “pretty”, “quantile”, “kmeans”, “hclust”, “bclust”, “fisher”, & “jenks”). For details, please see help, just type ?classIntervals in R console
First, we have to convert raster to vector using values function. Then, you have to create a object with a quantile interval applying classIntervals function to vector data.
[1] -8 7 20 35 49 65 87 117 164 267 985Now, we plot the DEM raster using the this custom break. We will use sppot function from sp package to plot raster data with state boundary shape file. Before that we will create a custom color palette using colorRampPalette of RColorBrewer.
rgb.palette <- colorRampPalette(c("lightblue","blue","green","yellow","red"),
space = "rgb")
#polys<- list("sp.lines", as(dem, "SpatialLines"), col="grey", lty=1) # CO state boundary
spplot(dem.BTM, main = "Quantile of DEM (m)",
at=at,
#sp.layout=list(polys),
par.settings=list(axis.line=list(col="darkgrey",lwd=1)),
colorkey=list(space="right",height=1, width=1.3,at=1:11,labels=list(cex=1.0,at=1:11,
labels=c("-8","7","20","35","49","65","87","117","164","267","985"))),
col.regions=rgb.palette(100))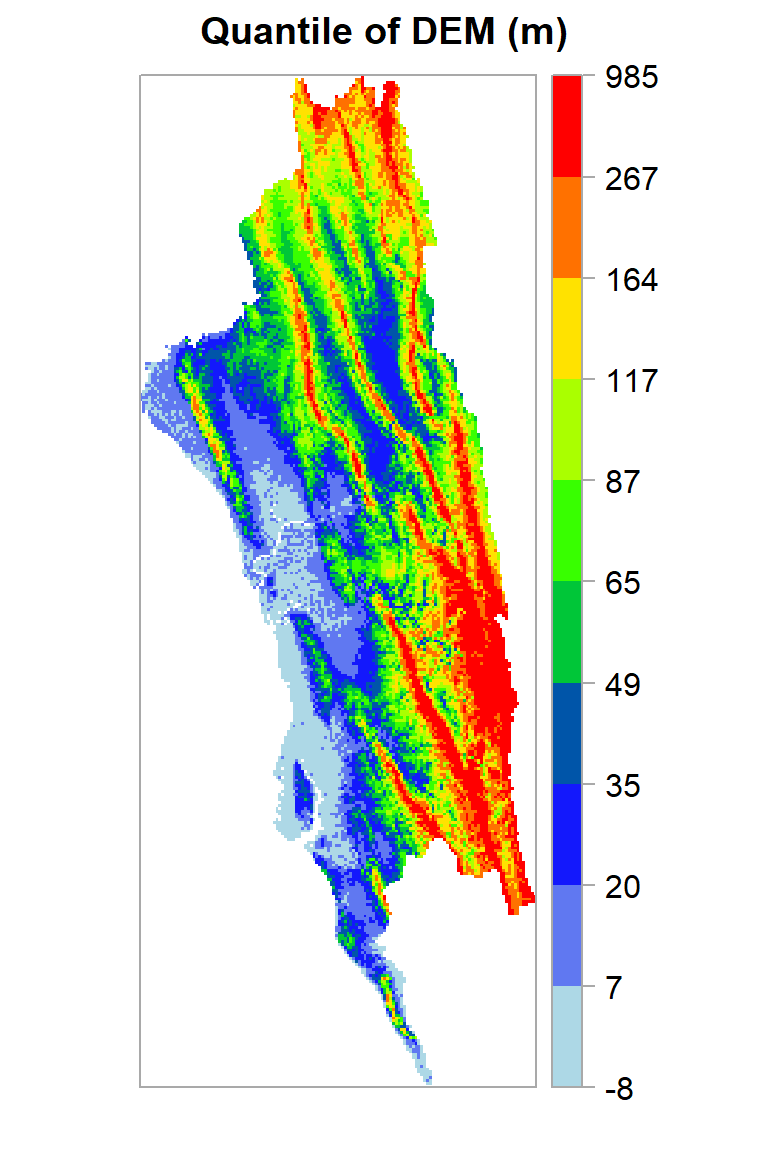
Focal statistics, also known as neighborhood or moving window statistics, involve performing calculations on a raster dataset using a moving window that slides over each cell. This window defines a local area around each cell, and calculations are carried out based on the values within that window. This technique is commonly used for various geospatial analysis tasks, such as smoothing, edge detection, and texture analysis.
In R, you can perform focal statistics using packages like raster and terra.
To illustrate the neighborhood processing for Focal Statistics calculating a Sum statistic, consider the processing cell with a value of 5 in the following diagram. A rectangular 3 by 3 cell neighborhood shape is specified. The sum of the values of the neighboring cells (3 + 2 + 3 + 4 + 2 + 1 + 4 = 19) plus the value of the processing cell (5) equals 24 (19 + 5 = 24). So a value of 24 is given to the cell in the output raster in the same location as the processing cell in the input raster.

The above figure demonstrates how the calculations are performed on a single cell in the input raster. In the following diagram, the results for all the input cells are shown. The cells highlighted in yellow identify the same processing cell and neighborhood as in the example above.
We will calculate focal mean of each cell of DEM data using focal() function of terra package.
Raster algebra, also known as map algebra, involves performing mathematical operations on raster datasets to create new raster outputs. It’s a fundamental technique in geospatial analysis that allows you to combine, modify, and analyze raster data using various arithmetic and logical operations.
A simple sample of raster algebra in R is to convert unit of DEM raster from meter to feet (1 m = 3.28084 feet)
Raster aggregation involves converting a high-resolution raster dataset into a lower-resolution raster dataset by grouping or summarizing cells into larger cells. This process reduces the level of detail in the data and can help improve processing efficiency and visualization for large datasets. Aggregation is often used when working with raster data at different scales, or when matching the resolution of one raster to another for analysis or visualization purposes.
First, we will create 500 x 500 km raster (mean & standard deviation) from 250 m x 250 m raster using aggregate function of raster package. To achieve this, we can combine pixels 2 by 2 (horizontally and vertically) using the aggregate() function and the argument fact = 2.
class : SpatRaster
dimensions : 665, 261, 1 (nrow, ncol, nlyr)
resolution : 500, 500 (x, y)
extent : 648050.4, 778550.4, 294767.3, 627267.3 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source(s) : memory
name : Band_1
min value : -4.500583
max value : 922.961380 If aggregating is the process of combining pixels, disaggregating is the process of splitting pixels into smaller ones. This operation is done with disagg(). Using the original 250m x 250m dem, each pixel will be disaggregated into smaller pixels (50 m x 50 m), once again using the fact argument.
class : SpatRaster
dimensions : 6645, 2605, 1 (nrow, ncol, nlyr)
resolution : 50, 50 (x, y)
extent : 648050.4, 778300.4, 295017.3, 627267.3 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source(s) : memory
name : Band_1
min value : -7.796429
max value : 985.049622 Resampling is a technique used in geospatial analysis to change the spatial resolution or alignment of raster datasets. Resampling involves calculating new pixel values based on the original pixel values within a defined neighborhood or using interpolation methods. It’s often used when you need to match the resolution of one raster to another, change the resolution of a raster for analysis, or prepare data for visualization.
We will re-sample 500 m raster to 250 raster again using dem (250 m). We will use resample() function. Here two methods available: bilinear for bilinear interpolation, or **ngb* for using the nearest neighbor. First argument is a raster object to be re-sampled and second argument is raster object with parameters that first raster should be re-sampled to.
class : SpatRaster
dimensions : 1329, 521, 1 (nrow, ncol, nlyr)
resolution : 250, 250 (x, y)
extent : 648050.4, 778300.4, 295017.3, 627267.3 (xmin, xmax, ymin, ymax)
coord. ref. : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source(s) : memory
name : Band_1
min value : -3.292471
max value : 908.095215 Raster mosaicking is the process of combining multiple raster datasets to create a single, continuous raster dataset that covers a larger geographic area. Mosaicking is commonly used when you have multiple raster images that need to be merged into a seamless and complete representation of a larger region. This technique is often employed in remote sensing, geospatial analysis, and cartography.
We will use merge() function to mosaic four raster in ./Raster/HILLY_DEM folder to create a seamless raster of hilly area of Bangladesh
[1] "G:\\My Drive\\Data\\Bangladesh\\.//Raster/HILLY_DEM/bandarban_dem_BTM.tiff"
[2] "G:\\My Drive\\Data\\Bangladesh\\.//Raster/HILLY_DEM/chittagong_dem_BTM.tiff"
[3] "G:\\My Drive\\Data\\Bangladesh\\.//Raster/HILLY_DEM/cox_bazar_dem_BTM.tiff"
[4] "G:\\My Drive\\Data\\Bangladesh\\.//Raster/HILLY_DEM/khagrachhari_dem_BTM.tiff"
[5] "G:\\My Drive\\Data\\Bangladesh\\.//Raster/HILLY_DEM/rangamati_dem_BTM.tiff" [[1]]
class : RasterLayer
dimensions : 520, 261, 135720 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 712800.4, 778050.4, 344017.3, 474017.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source : bandarban_dem_BTM.tiff
names : Band_1
values : -1.773466, 985.0496 (min, max)
[[2]]
class : RasterLayer
dimensions : 500, 323, 161500 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 648050.4, 728800.4, 418267.3, 543267.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source : chittagong_dem_BTM.tiff
names : Band_1
values : -4.444613, 279.4592 (min, max)
[[3]]
class : RasterLayer
dimensions : 523, 219, 114537 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 690300.4, 745050.4, 295017.3, 425767.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source : cox_bazar_dem_BTM.tiff
names : Band_1
values : -7.796429, 232.1743 (min, max)
[[4]]
class : RasterLayer
dimensions : 464, 191, 88624 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 675550.4, 723300.4, 510267.3, 626267.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source : khagrachhari_dem_BTM.tiff
names : Band_1
values : 13.54026, 477.2253 (min, max)
[[5]]
class : RasterLayer
dimensions : 834, 298, 248532 (nrow, ncol, ncell)
resolution : 250, 250 (x, y)
extent : 697800.4, 772300.4, 418767.3, 627267.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=-2000000 +datum=WGS84 +units=m +no_defs
source : rangamati_dem_BTM.tiff
names : Band_1
values : 10.69665, 963.0776 (min, max)For convert raster data to spatial point data frame, we will use rasterToPoints() function and then we will apply as.data.frame() function to convert a regular data frame
num [1:310665, 1:3] 720925 720175 720425 720675 720925 ...
- attr(*, "dimnames")=List of 2
..$ : NULL
..$ : chr [1:3] "x" "y" "Band_1" x y Band_1
1 720925.4 627142.3 357.0371
2 720175.4 626892.3 318.1637
3 720425.4 626892.3 320.6357
4 720675.4 626892.3 317.4960
5 720925.4 626892.3 355.9710
6 721175.4 626892.3 440.9489Converting vector data to raster data is a common operation in geospatial analysis, allowing you to represent vector features as pixel values in a raster grid. This conversion is often done when you need to integrate vector data with other raster datasets, perform raster-based analysis, or prepare data for modeling or visualization. In R, you can perform vector to raster conversion using packages like raster and terra. Here’s how you can use these packages to convert vector data to raster data:
We will create raster of district wise boro production in Bangladesh. We will use raster::rasterize() function to create this raster similar resoluation of DEM (250 m x 25 m)
class : RasterLayer
dimensions : 2680, 1919, 5142920 (nrow, ncol, ncell)
resolution : 249.9291, 249.3885 (x, y)
extent : 298487.8, 778101.8, 278578.1, 946939.2 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=0 +lon_0=90 +k=0.9996 +x_0=500000 +y_0=0 +datum=WGS84 +units=m +no_defs
source : memory
names : layer
values : 0, 1018.506 (min, max)Zonal statistics is a geospatial analysis technique that involves calculating summary statistics within predefined zones or regions of interest. These regions are typically defined by a vector dataset, such as polygons representing administrative boundaries, land parcels, or any other spatial units. Zonal statistics allow you to extract information from raster datasets based on the spatial boundaries defined by the vector features.
In this exercise we will calculate mean elevation five district of hilly area of Bangladesh.
We can apply raster::extract() function to calculate any statistics by zones
ID Band_1
1 1 192.32852
2 2 23.79157
3 3 17.54851
4 4 97.13356
5 5 142.51197 ID Band_1 dist.name
1 1 192.32852 Bandarban
2 2 23.79157 Chittagong
3 3 17.54851 Cox's Bazar
4 4 97.13356 Khagrachhari
5 5 142.51197 RangamatiYou can visualize the mean elevation using ggplot():
ggplot(as.data.frame(mean.elevation), aes(x=reorder(dist.name, +Band_1), y=Band_1)) +
geom_bar(stat="identity", position=position_dodge(),width=0.5, fill="gray") +
# add y-axis title and x-axis title leave blank
labs(y="Elevation (m))", x = "")+
# add plot title
ggtitle("Mean Elevation (m)")+
coord_flip()+
# customize plot themes
theme(
axis.line = element_line(colour = "gray"),
# plot title position at center
plot.title = element_text(hjust = 0.5),
# axis title font size
axis.title.x = element_text(size = 14),
# X and axis font size
axis.text.y=element_text(size=12,vjust = 0.5, hjust=0.5, colour='black'),
axis.text.x = element_text(size=12))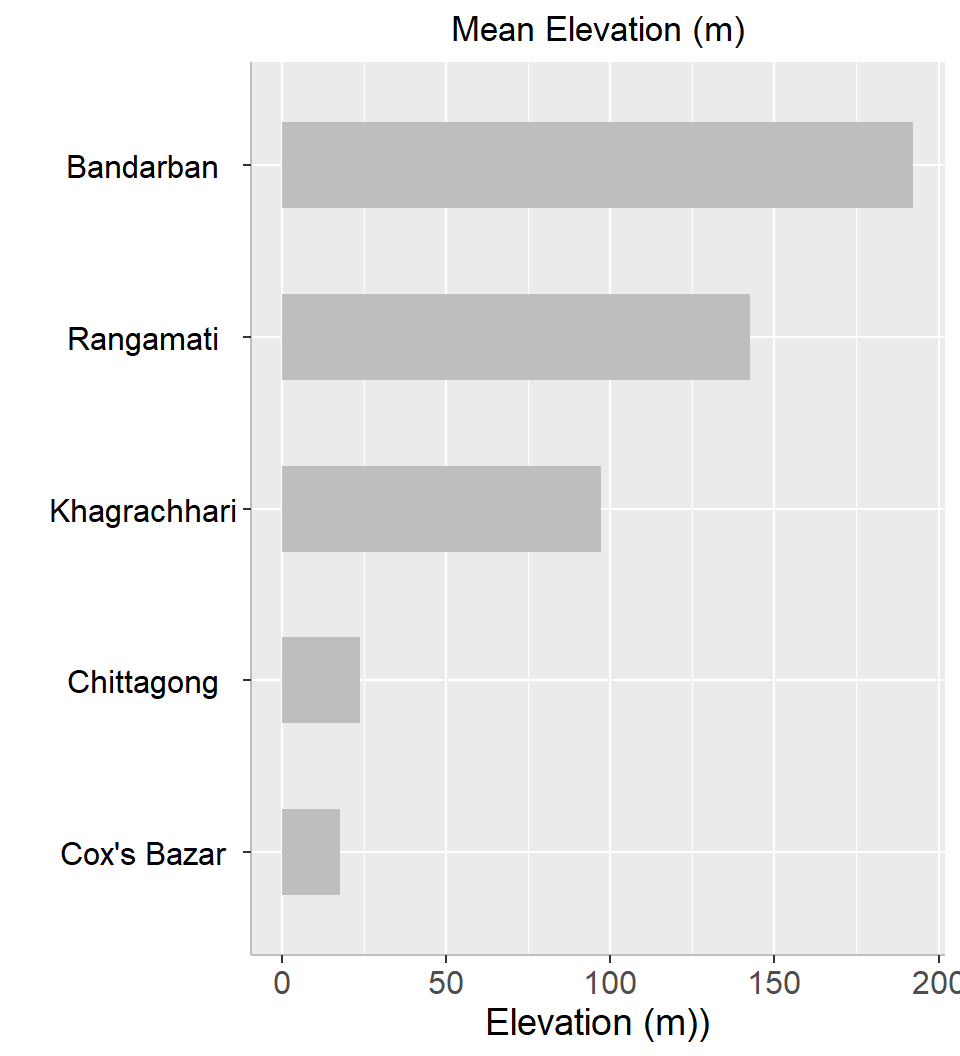
[{fig-align="left" width="200"}](https://github.com/zia207/r-colab/blob/main/working_with_raster_data.ipynb)
# Working with Raster Data in R {.unnumbered}
Raster data processing in R is commonly done using packages such as **`raster`** and **`rgdal`**. Raster data consists of grids of cells, where each cell holds a value representing some information, such as elevation, temperature, or land cover type. The **`raster`** package provides functions for reading, manipulating, and analyzing raster data, while the **`rgdal`** package focuses on reading and writing geospatial raster data formats.
The **`terra`** package is an advanced geospatial package in R that provides powerful tools for working with raster data. It is designed to handle large datasets efficiently and offers a wide range of capabilities for raster data processing, analysis, and visualization. **`terra`** is considered a successor to the **`raster`** package and aims to provide better performance and support for more complex operations.
Here are some key features and concepts of the **`terra`** package:
1. **Efficiency and Performance:** **`terra`** is optimized for performance, making it suitable for processing large raster datasets. It leverages modern computing hardware and parallel processing capabilities to efficiently handle computations.
2. **Raster Objects:** **`terra`** introduces the concept of "SpatioTemporalRaster" objects, which are used to represent raster data along with its spatial and temporal attributes.
3. **Basic Operations:** Similar to the **`raster`** package, **`terra`** allows you to perform basic operations like reading, writing, displaying, and extracting values from raster data.
4. **Spatial and Temporal Operations:** **`terra`** supports spatial and temporal operations such as cropping, reprojecting, resampling, masking, and aggregating raster data. It also handles multi-layer raster stacks efficiently.
5. **Advanced Analytics:** The package provides functions for more advanced spatial analytics, including focal operations (neighborhood analysis), zonal statistics, and mathematical operations on raster layers.
6. **Memory Management:** **`terra`** uses a "lazy" evaluation approach, which means that computations are performed only when needed. This allows for more memory-efficient processing of large datasets.
7. **Visualization:** The package includes visualization functions to help you create maps and plots of raster data.
8. **Integration with Other Packages:** **`terra`** can be integrated with other geospatial packages like **`sf`** for vector data handling and analysis.
In this exerciser we will learn following raster processing techniques with R:
- Basic Raster Operation
- Clipping
- Reclassification
- Focal Statistics
- Raster Algebra
- Aggregation
- Disaggregation
- Resample
- Mosaic
- Vector to raster conversion
- Convert Raster to Point Data
- Convert Point Data to Raster
- Raster Stack and Raster Brick
### Load R packages
```{r }
#| warning: false
#| error: false
library (terra)
library(sf)
library (rgdal)
library(tidyverse)
library(rgeos)
library(maptools)
library(classInt)
library(RColorBrewer)
library(raster)
```
### Data
We will use SRTM elevation data (250 m x 250 m) of all hilly districts of Bangladesh and The data can be found [here](https://github.com/zia207/r-colab/tree/main/Data/) for download.
We will use **rast()** function of **terra** package to load raster in R
```{r}
#| warning: false
#| error: false
dataFolder<-"G:\\My Drive\\Data\\Bangladesh\\"
# if data in working directory
dem<-terra::rast(paste0(dataFolder, "/Raster/hilly_DEM_UTM.tif"))
```
We can load data from github:
```{r}
#| warning: false
#| error: false
dem= terra::rast("/vsicurl/https://github.com//zia207/r-colab/raw/main/Data/Bangladesh/Raster/hilly_DEM_UTM.tif")
```
You can also load data with raster packages, but raster object relatively slow to perform some tasks. The terra library is more recent (but faster). We can convert a RasterLayer (i.e. a raster object) into a SpatRaster (i.e. a terra object):
```{r}
dem.r<-raster::raster(paste0(dataFolder, "/Raster/hilly_DEM_UTM.tif"))
dem.r
```
```{r}
# terra object
dem.t<-as(dem.r, "SpatRaster")
dem.t
```
### Basic Raster Operation
#### Check projection system
```{r}
#| warning: false
#| error: false
crs(dem, parse = TRUE)
```
#### Check raster attribute
```{r}
#| warning: false
#| error: false
dem
```
#### Raster extent
```{r}
ext(dem)
```
#### Raster statistics
R allow you to calculate some basic statistics of raster data. We can use **summary** function to get some statistics of raster layer.
```{r}
#| warning: false
#| error: false
summary(dem)
```
#### Histogram of raster
R-base function **hist()** is also useful to know frequency of raster values
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 4.5
HISTO.DEM<-hist(dem,
breaks=6,
col= "purple", # color of histogram
main="Distribution of Elevation values with 5 breaks", # tittle of the plot
xlab= "Elevation (m)", # x-axis name
ylab = "Number of Pixels") # y-axis name
box()
```
#### Plot raster
We can plot raster in diffrent ways in R. R base function **plot()** can easily apply on raster object for visualization:
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 7
plot(dem, main="DEM of Hilly District of Bangladesh")
```
R has an **image()** function that allows you to control the way a raster is rendered on the screen. The plot() function in R has a base setting for the number of pixels that it will plot (100,000 pixels). The image command thus might be better for rendering larger rasters.
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 7
image(dem)
```
To visualize raster in R using ggplot2, first we need to convert it to a dataframe. The terra package has an built-in function for conversion to a plotable dataframe. Then we can applly **geom_raster()** of ggplot().
```{r}
dem.df <- as.data.frame(dem, xy = TRUE)%>%
glimpse()
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 7
dem %>% as.data.frame(xy = TRUE) %>%
ggplot() +
geom_raster(aes(x = x, y = y, fill = Band_1)) +
scale_fill_viridis_c() +
coord_quickmap()
```
### Clipping
Raster clipping refers to the process of extracting a portion of a raster dataset based on a specified spatial extent or boundary. This is a common operation in geospatial analysis when you need to focus on a specific area of interest within a larger raster dataset. Raster clipping can help reduce the dataset's size, improve processing efficiency, and facilitate analysis that's relevant to a particular region.
In R, you can perform raster clipping using various packages, including **`raster`**, **`terra`**, and others. I'll provide examples using both the **`raster`** and **`terra`** packages.
In this exercise, we will clip hilly area DEM with extent of Bandarban district shape file. First we have to create a shape file of this district:
```{r}
bd.dist = sf::st_read("/vsicurl/https://github.com//zia207/r-colab/raw/main/Data/Bangladesh//Shapefiles//bd_district_BTM.shp")
```
```{r}
bandarban.dist = bd.dist %>%
rename(DIST_Name = ADM2_EN) %>%
filter(DIST_Name == "Bandarban")
```
Before clipping, you have to make sure both raster and vector data are in same projection systems.
```{r}
identical(crs(dem), crs(bandarban.dist ))
```
It looks like DEM in UTM and district shape files is BUTM project system. We have bring them in same projection system before clipping. Here we reproject DEM to BUTM using district shape files
```{r}
dem.BTM <- terra::project(dem, crs(bandarban.dist))
#terra::writeRaster(dem.BTM, paste0(dataFolder,"Raster/hilly_dem_BTM.tiff"), filetype = "GTiff", overwrite = TRUE)
```
```{r}
identical(crs(dem.BTM), crs(bandarban.dist ))
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 7
plot(dem.BTM)
plot(bandarban.dist$geometry, add=TRUE)
```
In R, clipping of a raster is two steps procedure, first you have to apply **crop()** function & then **mask** functions of **raster** package. **crop** returns a geographic subset of an object as specified by an **extent** object (or object from which an extent object can be extracted/created). **mask** Create a new Raster object that has the same values as input raster.
```{r}
#| warning: false
#| error: false
bandarban.dem_01<- crop(dem.BTM, ext(bandarban.dist))
bandarban.dem_02<-mask(bandarban.dem_01, bandarban.dist)
```
```{r}
#| warning: false
#| error: false
#| fig.width: 6
#| fig.height: 5
bandarban.dem_02 %>% as.data.frame(xy = TRUE) %>%
ggplot() +
geom_raster(aes(x = x, y = y, fill = Band_1)) +
scale_fill_viridis_c() +
coord_quickmap()
```
We can write raster using terra::writeRaster function:
```{r}
#| warning: false
#| error: false
terra::writeRaster(bandarban.dem_02, paste0(dataFolder,"Raster/bandarban_dem_BTM.tiff"), filetype = "GTiff", overwrite = TRUE)
#terra::writeRaster(bandarban.dem, "/content/drive/MyDrive/Data/Bangladesh/Raster/bandarban_dem_BTM.tiff",filetype = "GTiff", overwrite = TRUE )
```
### Reclassification
Raster reclassification is the process of assigning new values to the cells of a raster dataset based on predefined criteria. This operation is commonly used in geospatial analysis to reclassify continuous raster data (e.g., elevation, temperature) into discrete categories (e.g., low, medium, high).
In R, you can perform raster reclassification using packages like raster or terra packages
#### Histogram
Before reclassification, we will create a histogram with 5 breaks, of dem raster
```{r}
#| warning: false
#| error: false
#| fig.width: 6
#| fig.height: 5
hist.dem<-hist(dem.BTM,
breaks=4,
col= "purple", # color of histogram
main="Distribution of Elevation values with 5 breaks", # tittle of the plot
xlab= "Elevation (m)", # x-axis name
ylab = "Number of Pixels") # y-axis name
box()
```
Now, you can check where are breaks and how many pixels in each category, and use these breaks to plot a raster layer
```{r}
#| warning: false
#| error: false
hist.dem$breaks
hist.dem$counts
```
```{r}
#| warning: false
#| error: false
#| fig.width: 6
#| fig.height: 5
# plot using the histogram breaks.
plot(dem.BTM,
breaks = c(0, 200, 400, 600, 800, 1000),
col = terrain.colors(7),
main="DEM ")
```
We will use following breaks of histogram to reclassify the dem raster. First, you have to create classification matrix
- Class 1: -200 - 0 m (low elevation)
- Class 2: 0 - 400 m (medium elevation)
- Class 3: 400 - 1000 (Inf) m (high elevation)
```{r}
#| warning: false
#| error: false
# Create a dataframe
reclass_df <- c(-200, 200, 1,
200, 400, 2,
400, Inf, 3)
reclass_df
```
Next, you have to reshape the data frame to matrix using **matrix** function
```{r}
#| warning: false
#| error: false
reclass_m <- matrix(reclass_df,
ncol = 3,
byrow = TRUE)
reclass_m
```
Now you have to apply **terra::classify()** function of raster package using re-class_m object.
```{r}
#| warning: false
#| error: false
dem.reclass <- terra::classify(dem.BTM,
reclass_m)
```
You can view the distribution of pixels assigned to each class using the bar-plot().
```{r}
#| fig.width: 4
#| fig.height: 6
barplot(dem.reclass,
main = "Number of pixels in each class")
box()
```
Now we plot the reclassified raster using **plot** function with a nice looking legend
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
plot(dem.reclass,
legend = FALSE,
col = c("green", "blue", "red"), axes = FALSE,
main = "Reclassified DEM")
legend("topright",
legend = c("Low", "Medium", "High"),
fill = c("green", "blue", "red"),
border = FALSE,
bty = "n") # turn off legend border
box()
```
#### Quantile
If you want create a raster map using the quantile, you fave to use **classInt** package. The **classIntervals** function is create a custom breaks of raster values using different styles (such as "fixed", "sd", "equal", "pretty", "quantile", "kmeans", "hclust", "bclust", "fisher", & "jenks"). For details, please see help, just type **?classIntervals** in R console
First, we have to convert raster to vector using **values** function. Then, you have to create a object with a quantile interval applying **classIntervals** function to vector data.
```{r}
#| warning: false
#| error: false
value.vector <- values(dem.BTM)
value.vector <- value.vector[value.vector != 0]
at = round(classIntervals(value.vector, n = 10, style = "quantile")$brks, 0) # 10 quantile
at
```
Now, we plot the DEM raster using the this custom break. We will use **sppot** function from **sp** package to plot raster data with state boundary shape file. Before that we will create a custom color palette using **colorRampPalette** of **RColorBrewer**.
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
rgb.palette <- colorRampPalette(c("lightblue","blue","green","yellow","red"),
space = "rgb")
#polys<- list("sp.lines", as(dem, "SpatialLines"), col="grey", lty=1) # CO state boundary
spplot(dem.BTM, main = "Quantile of DEM (m)",
at=at,
#sp.layout=list(polys),
par.settings=list(axis.line=list(col="darkgrey",lwd=1)),
colorkey=list(space="right",height=1, width=1.3,at=1:11,labels=list(cex=1.0,at=1:11,
labels=c("-8","7","20","35","49","65","87","117","164","267","985"))),
col.regions=rgb.palette(100))
```
### Focal Statistics
Focal statistics, also known as neighborhood or moving window statistics, involve performing calculations on a raster dataset using a moving window that slides over each cell. This window defines a local area around each cell, and calculations are carried out based on the values within that window. This technique is commonly used for various geospatial analysis tasks, such as smoothing, edge detection, and texture analysis.
In R, you can perform focal statistics using packages like raster and terra.
To illustrate the neighborhood processing for Focal Statistics calculating a Sum statistic, consider the processing cell with a value of 5 in the following diagram. A rectangular 3 by 3 cell neighborhood shape is specified. The sum of the values of the neighboring cells (3 + 2 + 3 + 4 + 2 + 1 + 4 = 19) plus the value of the processing cell (5) equals 24 (19 + 5 = 24). So a value of 24 is given to the cell in the output raster in the same location as the processing cell in the input raster.
{width="433" height="189"}
The above figure demonstrates how the calculations are performed on a single cell in the input raster. In the following diagram, the results for all the input cells are shown. The cells highlighted in yellow identify the same processing cell and neighborhood as in the example above.
We will calculate focal mean of each cell of DEM data using **focal()** function of terra package.
```{r}
focal.sum<- focal(dem.BTM,
w=matrix(1/9,nrow=9,ncol=9), # matrix of weights for 9 x 9
fun=sum)
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
spplot(focal.sum, main="Focal Sum of DEM (m)")
```
```{r}
focal.mean<- focal(dem.BTM,
w=matrix(1/9,nrow=9,ncol=9), # matrix of weights for 3 x 3
fun=mean)
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
spplot(focal.mean,main="Focal Mean of DEM (m)")
```
### Raster Algebra
Raster algebra, also known as map algebra, involves performing mathematical operations on raster datasets to create new raster outputs. It's a fundamental technique in geospatial analysis that allows you to combine, modify, and analyze raster data using various arithmetic and logical operations.
A simple sample of raster algebra in R is to convert unit of DEM raster from meter to feet (1 m = 3.28084 feet)
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
dem.feet<-dem.BTM*3.28084
plot(dem.feet, main= "DEM (feet)")
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
dem.feet.1000<-dem.feet/1000
plot(dem.feet.1000, main = "DEM (feet x 1000)")
```
### Aggregation
Raster aggregation involves converting a high-resolution raster dataset into a lower-resolution raster dataset by grouping or summarizing cells into larger cells. This process reduces the level of detail in the data and can help improve processing efficiency and visualization for large datasets. Aggregation is often used when working with raster data at different scales, or when matching the resolution of one raster to another for analysis or visualization purposes.
First, we will create 500 x 500 km raster (mean & standard deviation) from 250 m x 250 m raster using **aggregate** function of **raster** package. To achieve this, we can combine pixels 2 by 2 (horizontally and vertically) using the aggregate() function and the argument fact = 2.
```{r}
#rm(mean)
#rm(sd)
mean.DEM<-aggregate(dem.BTM, fact=2, fun=mean)
sd.DEM<-aggregate(dem, fact=2, fun=sd)
se.DEM<-sd.DEM/sqrt(2)
mean.DEM
```
```{r}
#| warning: false
#| error: false
p1<-spplot(mean.DEM, main="500 m x 500 m of DEM")
p2<-spplot(sd.DEM, main="SD (m) ")
p3<-spplot(se.DEM, main="SE (m) )")
```
```{r}
#| warning: false
#| error: false
#| fig.width: 10
#| fig.height: 5
library(gridExtra)
grid.arrange(p1, p2, p3, ncol=3)
```
### Disaggregation
If aggregating is the process of combining pixels, disaggregating is the process of splitting pixels into smaller ones. This operation is done with **disagg()**. Using the original 250m x 250m dem, each pixel will be disaggregated into smaller pixels (50 m x 50 m), once again using the **fact** argument.
```{r}
dem.50 <- disagg(dem.BTM, fact = 5)
dem.50
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
spplot(dem.50, main="50 m x 50 m of DEM")
```
### Resampling
Resampling is a technique used in geospatial analysis to change the spatial resolution or alignment of raster datasets. Resampling involves calculating new pixel values based on the original pixel values within a defined neighborhood or using interpolation methods. It's often used when you need to match the resolution of one raster to another, change the resolution of a raster for analysis, or prepare data for visualization.
We will re-sample 500 m raster to 250 raster again using dem (250 m). We will use **resample()** function. Here two methods available: **bilinear** for bilinear interpolation, or **ngb* for using the nearest neighbor. First argument is a raster object to be re-sampled and second argument is raster object with parameters that first raster should be re-sampled to.
```{r}
dem.250m<-resample(mean.DEM, dem.BTM, method='bilinear') #
dem.250m
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
spplot(dem.250m, main="Resampled DEM - 250 m x 250 m")
```
### Mosaic
Raster mosaicking is the process of combining multiple raster datasets to create a single, continuous raster dataset that covers a larger geographic area. Mosaicking is commonly used when you have multiple raster images that need to be merged into a seamless and complete representation of a larger region. This technique is often employed in remote sensing, geospatial analysis, and cartography.
We will use *merge()* function to mosaic four raster in ./Raster/HILLY_DEM folder to create a seamless raster of hilly area of Bangladesh
```{r}
#| warning: false
#| error: false
DEM.input <- list.files(path= paste0(dataFolder,".//Raster/HILLY_DEM"),pattern=".tiff$",full.names=T)
DEM.input
```
```{r}
#| warning: false
#| error: false
r <- lapply(DEM.input, raster::raster)
r
```
```{r}
r1<-do.call(merge, c(r, tolerance = 1))
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 6
spplot(r1, main="Mosaic DEM (m)")
```
### Raster to vector conversion
For convert raster data to spatial point data frame, we will use **rasterToPoints()** function and then we will apply **as.data.frame()** function to convert a regular data frame
```{r}
dataFolder<-"G:\\My Drive\\Data\\Bangladesh\\"
# if data in working directory
r<-raster(paste0(dataFolder, "/Raster/hilly_DEM_BTM.tiff"))
```
```{r}
SPDF <- rasterToPoints(r)
str(SPDF)
df<-as.data.frame(SPDF)
head(df)
#write.csv(df, paste0(dataFolder,"GP_ElEV.csv", row.names = F))
```
### Vector to raster conversion
Converting vector data to raster data is a common operation in geospatial analysis, allowing you to represent vector features as pixel values in a raster grid. This conversion is often done when you need to integrate vector data with other raster datasets, perform raster-based analysis, or prepare data for modeling or visualization. In R, you can perform vector to raster conversion using packages like raster and terra. Here's how you can use these packages to convert vector data to raster data:
We will create raster of district wise boro production in Bangladesh. We will use **raster::rasterize()** function to create this raster similar resoluation of DEM (250 m x 25 m)
```{r}
#| warning: false
#| error: false
dataFolder<-"G:\\My Drive\\Data\\Bangladesh\\"
# if data in working directory
poly<-raster::shapefile(paste0(dataFolder, "Shapefiles/bd_district_rice_production_2018_2019_BTM.shp"))
bd.dem<-raster::raster(paste0(dataFolder, "Raster/DEM.tif"))
```
```{r}
poly$Boro_1000<-poly$Boro/1000
```
```{r}
#| warning: false
#| error: false
# Define the extent, resolution, and target raster template
r <- raster::raster(bd.dem)
extent(r) <- raster::extent(poly)
rp <- raster::rasterize(poly, r, 'Boro_1000')
rp
```
```{r}
#| warning: false
#| error: false
#| fig.width: 5
#| fig.height: 7
spplot(rp, main = "Boro Production Raster (MT X 1000)")
```
### Zoal Statistics
Zonal statistics is a geospatial analysis technique that involves calculating summary statistics within predefined zones or regions of interest. These regions are typically defined by a vector dataset, such as polygons representing administrative boundaries, land parcels, or any other spatial units. Zonal statistics allow you to extract information from raster datasets based on the spatial boundaries defined by the vector features.
In this exercise we will calculate mean elevation five district of hilly area of Bangladesh.
```{r}
dataFolder<-"G:\\My Drive\\Data\\Bangladesh\\"
# if data in working directory
hilly.dist<-raster::shapefile(paste0(dataFolder, "Shapefiles/hilly_district_BTM.shp"))
hilly.dem<-raster::raster(paste0(dataFolder, "Raster/hilly_dem_BTM.tiff"))
```
We can apply **raster::extract()** function to calculate any statistics by zones
```{r}
mean.elevation <- extract(hilly.dem, hilly.dist, fun=mean, na.rm=TRUE, df=TRUE)
mean.elevation
```
```{r}
# add district name
mean.elevation$dist.name<-levels(as.factor(hilly.dist$ADM2_EN))
mean.elevation
```
You can visualize the mean elevation using ggplot():
```{r}
#| warning: false
#| error: false
#| fig.width: 5
#| fig.height: 5.5
ggplot(as.data.frame(mean.elevation), aes(x=reorder(dist.name, +Band_1), y=Band_1)) +
geom_bar(stat="identity", position=position_dodge(),width=0.5, fill="gray") +
# add y-axis title and x-axis title leave blank
labs(y="Elevation (m))", x = "")+
# add plot title
ggtitle("Mean Elevation (m)")+
coord_flip()+
# customize plot themes
theme(
axis.line = element_line(colour = "gray"),
# plot title position at center
plot.title = element_text(hjust = 0.5),
# axis title font size
axis.title.x = element_text(size = 14),
# X and axis font size
axis.text.y=element_text(size=12,vjust = 0.5, hjust=0.5, colour='black'),
axis.text.x = element_text(size=12))
```
### Further Reading
1. [Raster data manipulation](https://rspatial.org/spatial/8-rastermanip.html#google_vignette)
2. [Raster Data in R - The Basics](https://www.neonscience.org/resources/learning-hub/tutorials/raster-data-r)
3. [Intro to Raster Data](https://datacarpentry.org/r-raster-vector-geospatial/01-raster-structure)