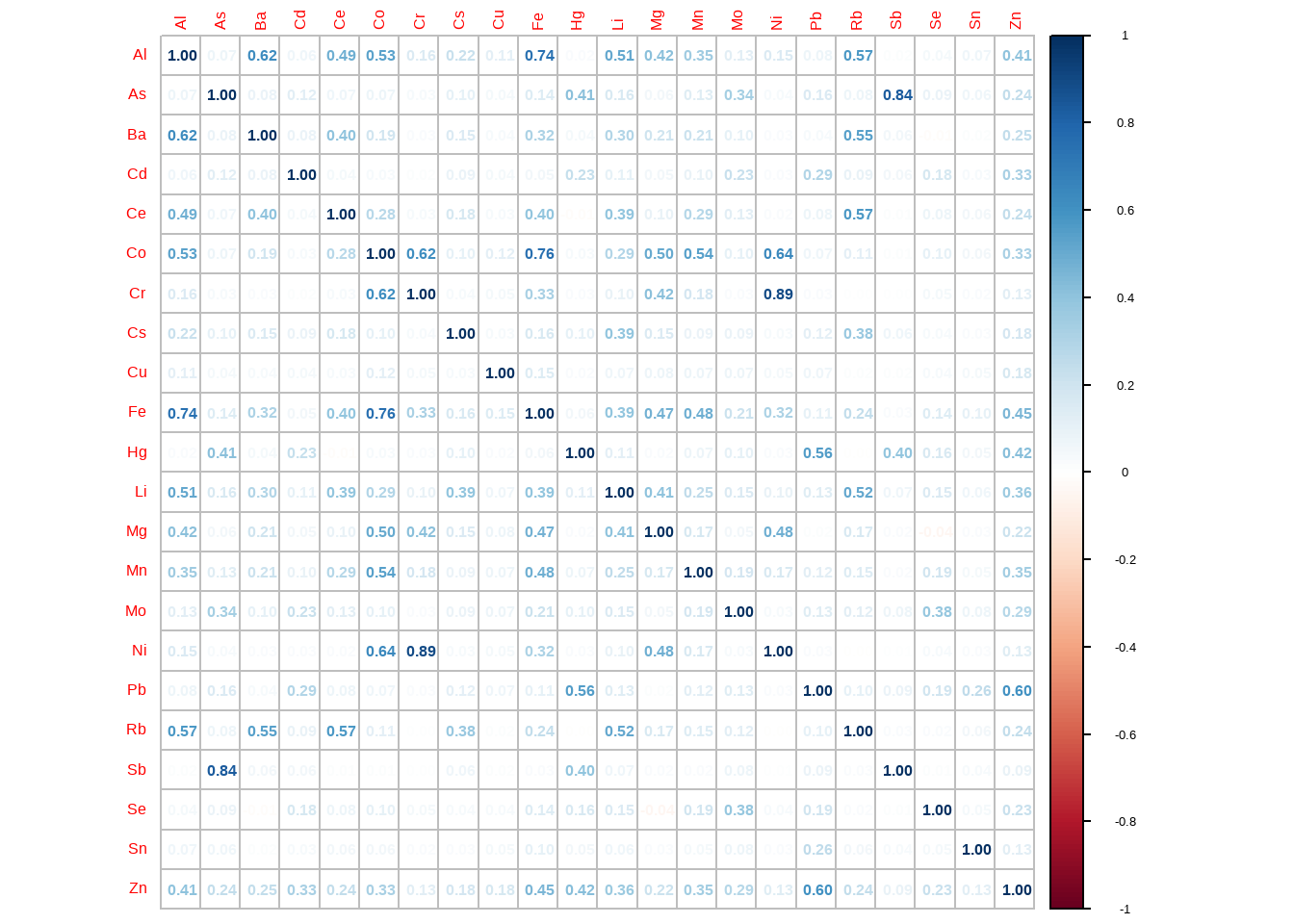
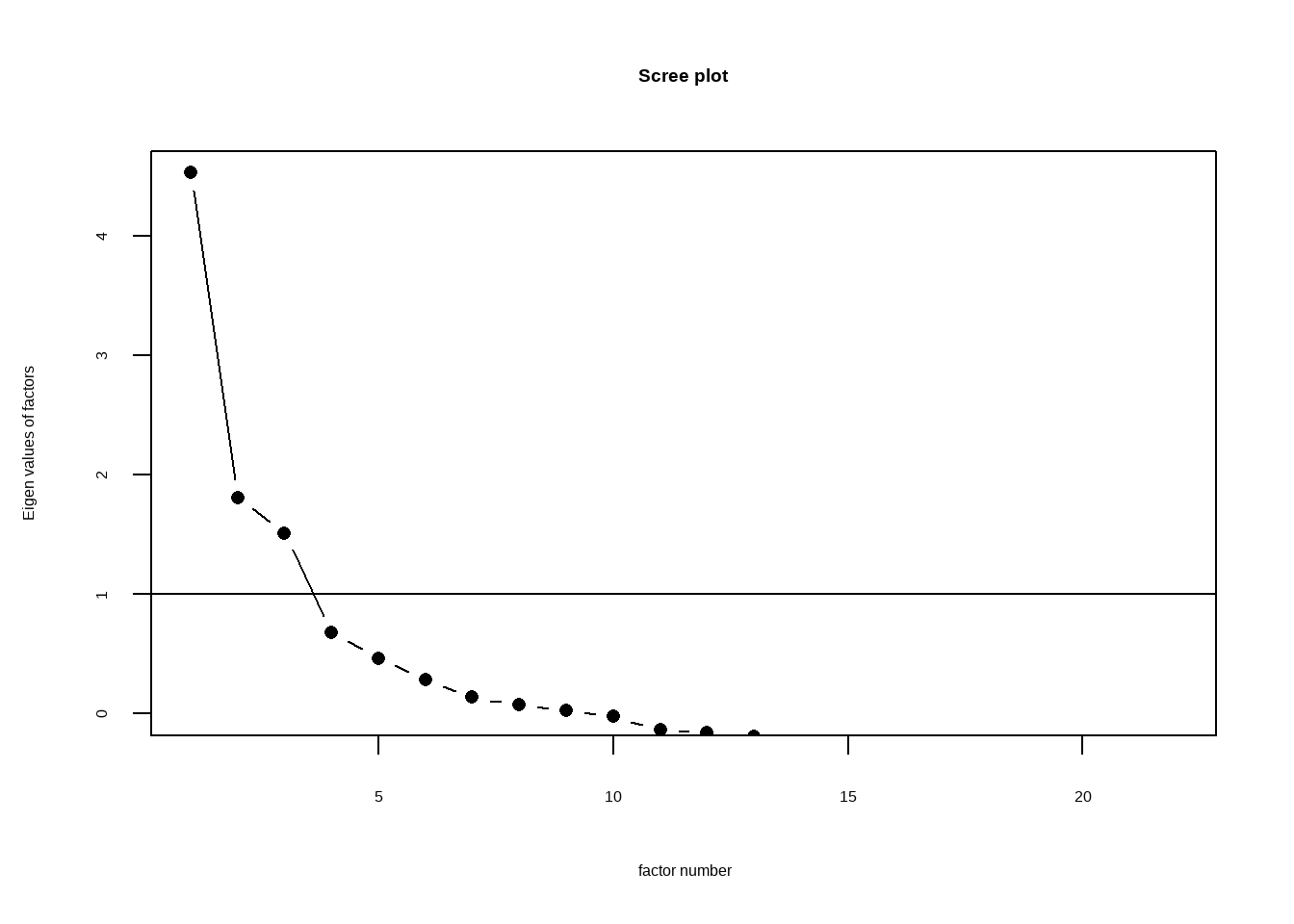
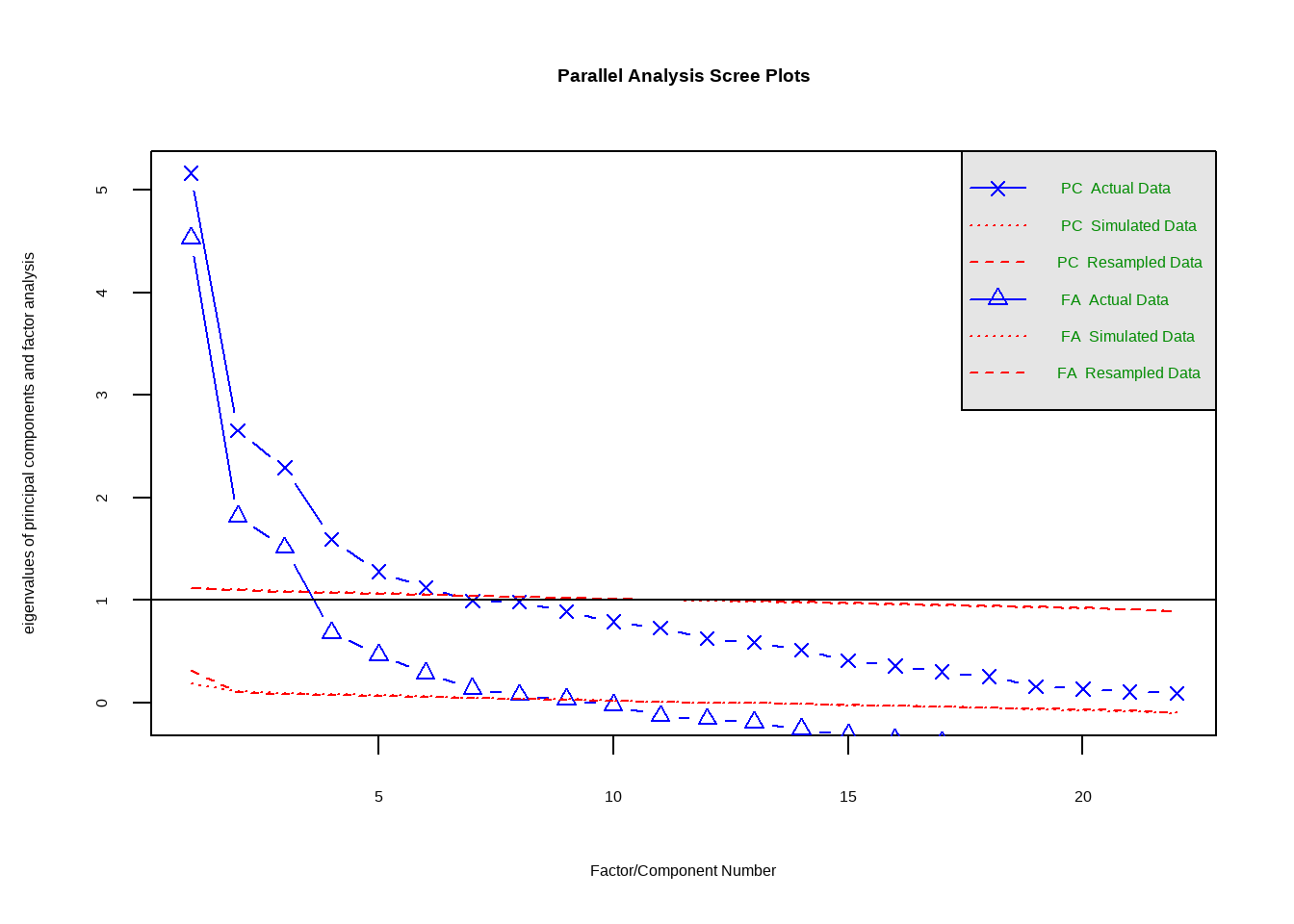
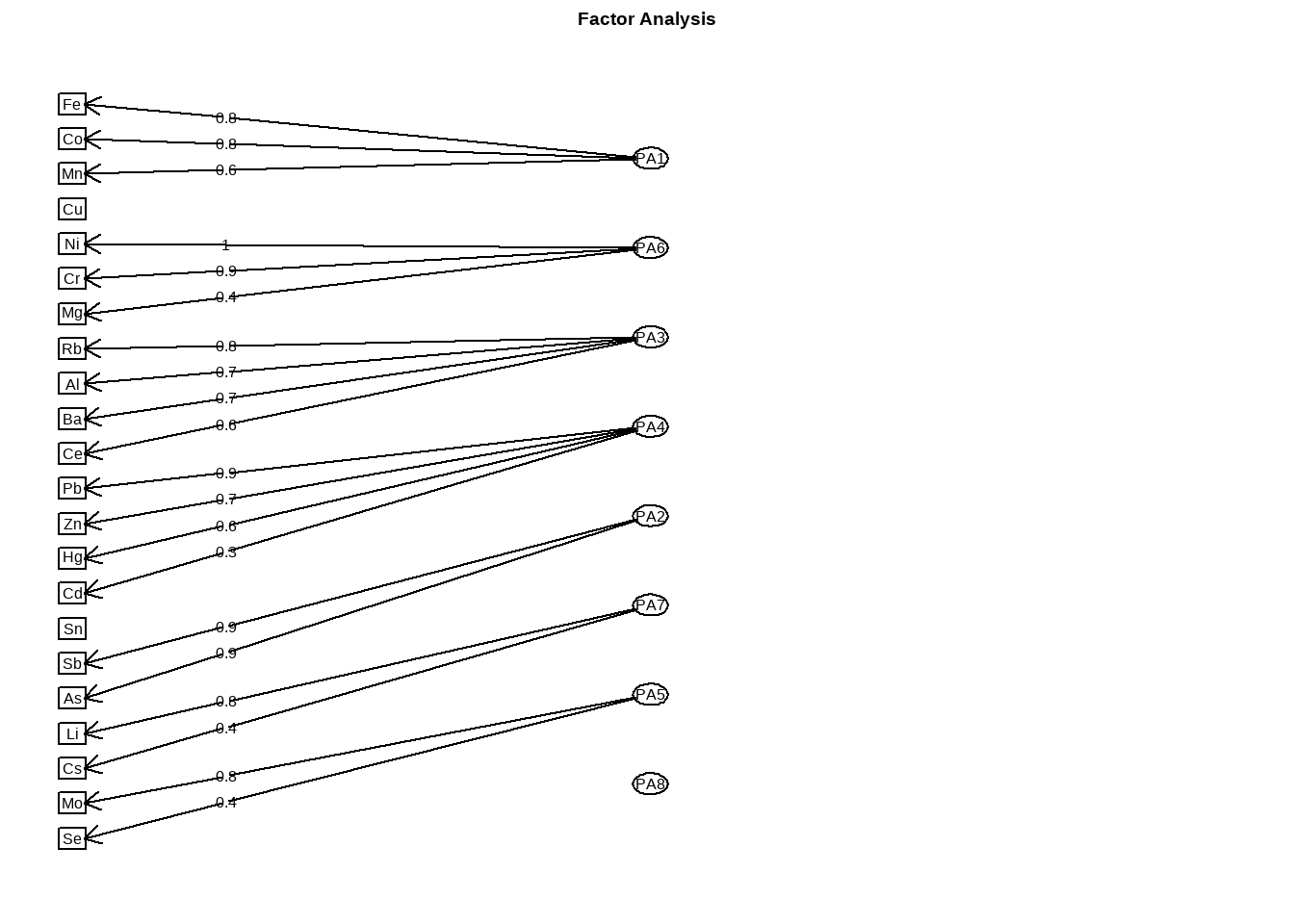
Rows: 4,813
Columns: 28
$ A_LabID <chr> "C-320500", "C-299963", "C-319572", "C-355112", "C-319622", …
$ SiteID <dbl> 847, 7209, 6811, 4072, 4315, 12782, 10830, 252, 1964, 6920, …
$ StateID <chr> "NV", "ME", "CO", "WI", "CO", "MT", "WA", "PA", "IN", "MO", …
$ Latitude <dbl> 38.8486, 44.2417, 37.8284, 42.7492, 39.2340, 46.3279, 46.499…
$ Longitude <dbl> -117.5824, -70.4833, -107.6216, -90.2205, -106.2517, -112.90…
$ LandCover <chr> "Shrubland", "Forested Upland", "Shrubland", "Forested Uplan…
$ Al <dbl> 7.43, 5.35, 5.82, 4.61, 6.24, 3.76, 6.99, 7.53, 9.06, 3.02, …
$ As <dbl> 195.0, 2.7, 18.4, 7.7, 25.1, 565.0, 4.2, 10.5, 94.5, 9.0, 5.…
$ Ba <dbl> 496, 352, 686, 610, 1030, 399, 706, 466, 414, 317, 372, 654,…
$ Cd <dbl> 16.1, 0.5, 5.1, 0.6, 3.2, 2.6, 0.6, 0.2, 1.4, 1.4, 1.1, 1.2,…
$ Ce <dbl> 42.0, 43.0, 67.9, 51.9, 48.9, 28.0, 67.9, 75.5, 104.0, 47.6,…
$ Co <dbl> 11.1, 5.2, 14.4, 9.5, 6.8, 8.9, 13.3, 21.1, 56.8, 11.7, 4.8,…
$ Cr <dbl> 42, 63, 24, 38, 9, 20, 48, 81, 217, 56, 24, 16, 59, 13, 54, …
$ Cs <dbl> 26.0, 2.5, 12.0, 2.5, 2.5, 5.0, 5.0, 2.5, 7.0, 2.5, 2.5, 2.5…
$ Cu <dbl> 87.9, 16.5, 141.0, 35.4, 147.0, 232.0, 34.7, 79.8, 98.7, 13.…
$ Fe <dbl> 4.17, 1.59, 3.63, 2.26, 7.39, 13.00, 3.44, 5.29, 8.28, 2.40,…
$ Hg <dbl> 8.24, 0.04, 0.07, 0.04, 1.32, 0.44, 0.03, 0.08, 0.32, 0.06, …
$ Li <dbl> 128, 23, 51, 19, 11, 28, 24, 37, 71, 16, 14, 57, 29, 29, 23,…
$ Mg <dbl> 1.62, 0.31, 0.77, 0.57, 0.34, 0.62, 0.80, 0.65, 0.39, 0.16, …
$ Mn <dbl> 740, 635, 4040, 719, 812, 6580, 713, 879, 480, 894, 430, 233…
$ Mo <dbl> 9.16, 1.06, 9.41, 0.87, 4.23, 70.30, 0.72, 2.10, 5.02, 1.33,…
$ Ni <dbl> 25.4, 34.8, 9.8, 20.3, 4.0, 7.2, 21.1, 31.9, 217.0, 11.1, 9.…
$ Pb <dbl> 2200.0, 34.0, 865.0, 49.1, 1510.0, 62.5, 83.0, 268.0, 259.0,…
$ Rb <dbl> 110.0, 79.0, 143.0, 72.5, 89.1, 46.6, 64.7, 88.6, 69.8, 51.6…
$ Sb <dbl> 32.30, 1.60, 6.04, 1.21, 6.26, 20.80, 0.77, 1.19, 9.84, 1.13…
$ Se <dbl> 3.1, 0.2, 0.4, 0.5, 0.7, 0.4, 0.1, 0.9, 1.3, 0.6, 0.4, 0.1, …
$ Sn <dbl> 3.2, 2.5, 2.1, 1.3, 66.9, 32.6, 2.0, 2.5, 82.4, 1.8, 9.7, 5.…
$ Zn <dbl> 2130, 1440, 977, 943, 770, 689, 612, 541, 536, 518, 491, 491…