Code
library(tidyverse)
library(tidymodels)
library(tensorflow)
library(keras)
use_virtualenv("r-reticulate")
Keras is a popular high-level deep learning library that provides a convenient and user-friendly API for building and training deep learning models. TensorFlow is a powerful open-source deep learning framework that serves as the backend for Keras and provides efficient computation for training and running deep learning models.
While Keras is primarily known for its Python implementation, it also has support for other programming languages, including R. In R, you can use the keras package which provides an interface to the Keras library with TensorFlow
To install TensorFlow in R, you can use the reticulate package, which provides an interface to Python from within R. Prior to using the tensorflow R package you need to install a version of Python and TensorFlow on your system. Below we describe how to install to do this as well the various options available for customizing your installation. Please follow instruction describe here.
library(reticulate)
path_to_python <- install_python()
virtualenv_create(“r-reticulate”, python = path_to_python)
install.packages(“keras”)
library(keras)
install_keras(envname = “r-reticulate”)
library(tensorflow)
tf$constant(“Hello Tensorflow!”)
In this exercise we will use following synthetic data set and will use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG variables to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
Rows: 1,408
Columns: 9
$ SOC <dbl> 1.900, 2.644, 0.800, 0.736, 15.641, 8.818, 3.782, 6.641, 4.803, …
$ DEM <dbl> 2825.1111, 2535.1086, 1716.3300, 1649.8933, 2675.3113, 2581.4839…
$ Slope <dbl> 18.981682, 14.182393, 1.585145, 9.399726, 12.569353, 6.358553, 1…
$ TPI <dbl> -0.91606224, -0.15259802, -0.39078590, -2.54008722, 7.40076303, …
$ MAT <dbl> 4.709227, 4.648000, 6.360833, 10.265385, 2.798550, 6.358550, 7.0…
$ MAP <dbl> 613.6979, 597.7912, 201.5091, 298.2608, 827.4680, 679.1392, 508.…
$ NDVI <dbl> 0.6845260, 0.7557631, 0.2215059, 0.2785148, 0.7337426, 0.7017139…
$ NLCD <chr> "Forest", "Forest", "Shrubland", "Shrubland", "Forest", "Forest"…
$ FRG <chr> "Fire Regime Group IV", "Fire Regime Group IV", "Fire Regime Gro…Review the joint distribution of a few pairs of columns from the training set using GGally package.
Rows: 1,408
Columns: 7
$ SOC <dbl> 1.900, 2.644, 0.800, 0.736, 15.641, 8.818, 3.782, 6.641, 4.803, …
$ DEM <dbl> 2825.1111, 2535.1086, 1716.3300, 1649.8933, 2675.3113, 2581.4839…
$ Slope <dbl> 18.981682, 14.182393, 1.585145, 9.399726, 12.569353, 6.358553, 1…
$ TPI <dbl> -0.91606224, -0.15259802, -0.39078590, -2.54008722, 7.40076303, …
$ MAT <dbl> 4.709227, 4.648000, 6.360833, 10.265385, 2.798550, 6.358550, 7.0…
$ MAP <dbl> 613.6979, 597.7912, 201.5091, 298.2608, 827.4680, 679.1392, 508.…
$ NDVI <dbl> 0.6845260, 0.7557631, 0.2215059, 0.2785148, 0.7337426, 0.7017139…Rows: 1,408
Columns: 15
$ DEM <dbl> 2825.1111, 2535.1086, 1716.3300, 1649.8933, …
$ Slope <dbl> 18.981682, 14.182393, 1.585145, 9.399726, 12…
$ TPI <dbl> -0.91606224, -0.15259802, -0.39078590, -2.54…
$ MAT <dbl> 4.709227, 4.648000, 6.360833, 10.265385, 2.7…
$ MAP <dbl> 613.6979, 597.7912, 201.5091, 298.2608, 827.…
$ NDVI <dbl> 0.6845260, 0.7557631, 0.2215059, 0.2785148, …
$ SOC <dbl> 1.900, 2.644, 0.800, 0.736, 15.641, 8.818, 3…
$ NLCD_Herbaceous <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0,…
$ NLCD_Planted.Cultivated <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ NLCD_Shrubland <dbl> 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ FRG_Fire.Regime.Group.II <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0,…
$ FRG_Fire.Regime.Group.III <dbl> 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0,…
$ FRG_Fire.Regime.Group.IV <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1,…
$ FRG_Fire.Regime.Group.V <dbl> 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ FRG_Indeterminate.FRG <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…Separate the target or response value—the “label” (SOC) from the features. This label is the value that you will train the model to predict.
It is good practice to normalize features that use different scales and ranges.
One reason this is important is because the features are multiplied by the model weights. So, the scale of the outputs and the scale of the gradients are affected by the scale of the inputs.
Although a model might converge without feature normalization, normalization makes training much more stable.
Note: There is no advantage to normalizing the one-hot features—it is done here for simplicity. For more details on how to use the preprocessing layers, refer to the Working with preprocessing layers guide and the Classify structured data using Keras preprocessing layers tutorial.
Source:(https://tensorflow.rstudio.com/tutorials/keras/regression#inspect-the-data
The layer_normalization() is a clean and simple way to add feature normalization into your model.
The first step is to create the layer:
Then, fit the state of the preprocessing layer to the data by calling adapt():
The normalization layer for a multiple-input model).
Two hidden, non-linear, Dense layers with the ReLU (relu) activation function nonlinearity.
A linear Dense single-output layer.
Both models will use the same training procedure so the compile method is included in the build_and_compile_model function below.
Model: "sequential"
________________________________________________________________________________
Layer (type) Output Shape Param # Trainable
================================================================================
normalization (Normalization) (None, 14) 29 Y
dense_2 (Dense) (None, 64) 960 Y
dense_1 (Dense) (None, 64) 4160 Y
dense (Dense) (None, 1) 65 Y
================================================================================
Total params: 5,214
Trainable params: 5,185
Non-trainable params: 29
________________________________________________________________________________Use Keras fit() to execute the training for 100 epochs:
[1] "RMSE: 3.5"[1] "MAE: 2.08"library(ggpmisc)
formula<-y~x
ggplot(test_labels, aes(SOC,Pred.SOC$V1)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("DNN-Keras") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))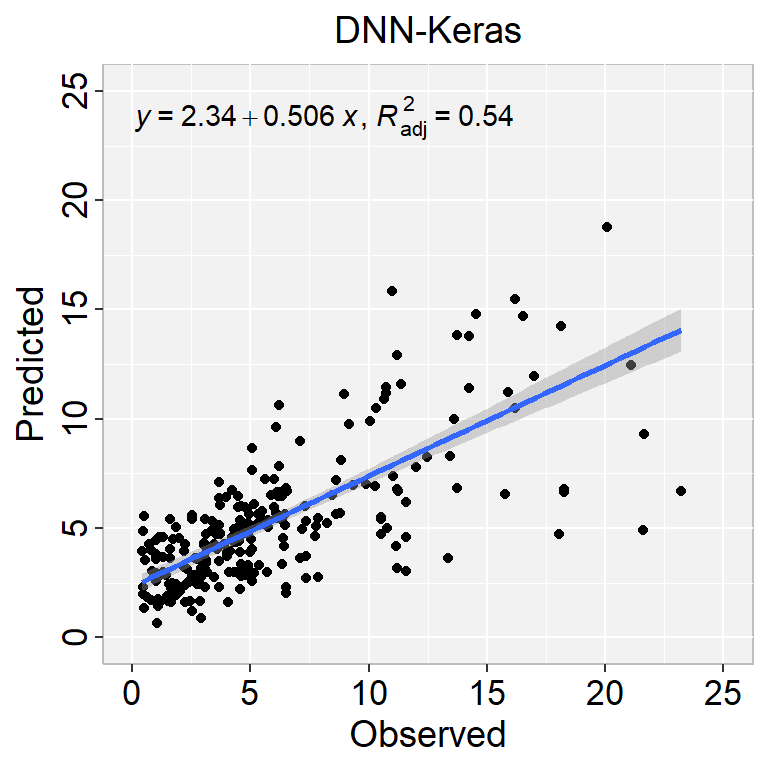
In this exercise we will use following synthetic data set and will use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG variables to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
Rows: 1,408
Columns: 9
$ SOC <dbl> 1.900, 2.644, 0.800, 0.736, 15.641, 8.818, 3.782, 6.641, 4.803, …
$ DEM <dbl> 2825.1111, 2535.1086, 1716.3300, 1649.8933, 2675.3113, 2581.4839…
$ Slope <dbl> 18.981682, 14.182393, 1.585145, 9.399726, 12.569353, 6.358553, 1…
$ TPI <dbl> -0.91606224, -0.15259802, -0.39078590, -2.54008722, 7.40076303, …
$ MAT <dbl> 4.709227, 4.648000, 6.360833, 10.265385, 2.798550, 6.358550, 7.0…
$ MAP <dbl> 613.6979, 597.7912, 201.5091, 298.2608, 827.4680, 679.1392, 508.…
$ NDVI <dbl> 0.6845260, 0.7557631, 0.2215059, 0.2785148, 0.7337426, 0.7017139…
$ NLCD <chr> "Forest", "Forest", "Shrubland", "Shrubland", "Forest", "Forest"…
$ FRG <chr> "Fire Regime Group IV", "Fire Regime Group IV", "Fire Regime Gro…The data set (n = 1408) will randomly split into sub-sets for training (70%), validation (15%) and test data (15%). The validation data will be used to optimized the model parameters during the tuning and training processes. The test data set will be used as the hold-out data to evaluate the DNN model.
dnn_recipe <-
recipe(SOC ~ ., data = train) %>%
step_zv(all_predictors()) %>%
step_dummy(all_nominal()) %>%
step_normalize(all_numeric_predictors())
# For testing when we arrive at a final model:
test_normalized <- bake(prep(dnn_recipe), new_data = test, all_predictors())
# Set 10 fold cross-validation data set
cv_folds<- vfold_cv(train, v=5)Single Layer Neural Network Model Specification (regression)
Main Arguments:
hidden_units = tune()
dropout = tune()
epochs = tune()
activation = relu
Engine-Specific Arguments:
verbose = 0
optimizer = optimizer_adam(0.001)
validation = 0.1
Computational engine: keras ══ Workflow ════════════════════════════════════════════════════════════════════
Preprocessor: Recipe
Model: mlp()
── Preprocessor ────────────────────────────────────────────────────────────────
3 Recipe Steps
• step_zv()
• step_dummy()
• step_normalize()
── Model ───────────────────────────────────────────────────────────────────────
Single Layer Neural Network Model Specification (regression)
Main Arguments:
hidden_units = tune()
dropout = tune()
epochs = tune()
activation = relu
Engine-Specific Arguments:
verbose = 0
optimizer = optimizer_adam(0.001)
validation = 0.1
Computational engine: keras # A tibble: 30 × 9
hidden_units dropout epochs .metric .estimator mean n std_err .config
<int> <dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 1 0.000795 125 mae standard 3.59 5 0.253 Preproce…
2 1 0.000795 125 rmse standard 5.49 5 0.461 Preproce…
3 1 0.000795 125 rsq standard 0.182 2 0.170 Preproce…
4 10 0.515 557 mae standard 2.84 5 0.0727 Preproce…
5 10 0.515 557 rmse standard 4.06 5 0.151 Preproce…
6 10 0.515 557 rsq standard 0.353 5 0.00975 Preproce…
7 9 0.414 345 mae standard 2.75 5 0.103 Preproce…
8 9 0.414 345 rmse standard 3.92 5 0.181 Preproce…
9 9 0.414 345 rsq standard 0.401 5 0.0208 Preproce…
10 5 0.0478 811 mae standard 2.76 5 0.129 Preproce…
# ℹ 20 more rowsSingle Layer Neural Network Model Specification (regression)
Main Arguments:
hidden_units = 5
dropout = 0.0477566176559776
epochs = 811
activation = relu
Engine-Specific Arguments:
verbose = 0
optimizer = optimizer_adam(0.001)
validation = 0.1
Computational engine: keras library(ggpmisc)
formula<-y~x
ggplot(test, aes(SOC,SOC.pred$.pred)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("DNN-Keras") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))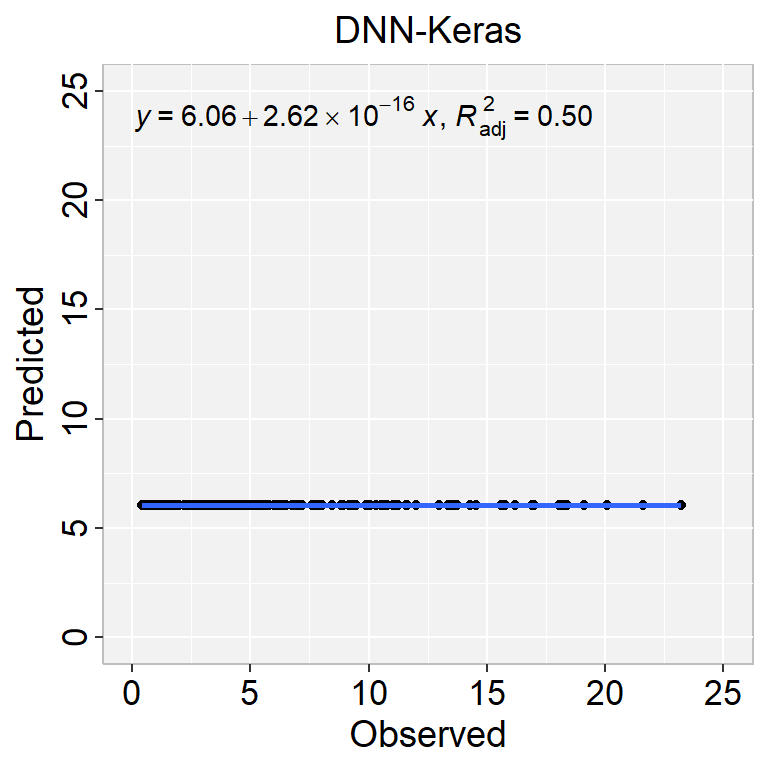
[{fig-align="left" width="268"}](https://github.com/zia207/r-colab/blob/main/Keras_R_colab.ipynb)
# Deep Neural Network with Keras-Tensorflow {.unnumbered}
{width="294"}
[Keras](https://keras.io/) is a popular high-level deep learning library that provides a convenient and user-friendly API for building and training deep learning models. [TensorFlow](https://www.tensorflow.org/) is a powerful open-source deep learning framework that serves as the backend for Keras and provides efficient computation for training and running deep learning models.
While Keras is primarily known for its Python implementation, it also has support for other programming languages, including R. In R, you can use the [keras package](https://posit.co/blog/keras-for-r/) which provides an interface to the Keras library with [TensorFlow](https://tensorflow.rstudio.com/)
### Installation
To install TensorFlow in R, you can use the **`reticulate`** package, which provides an interface to Python from within R. Prior to using the **tensorflow** R package you need to install a version of Python and TensorFlow on your system. Below we describe how to install to do this as well the various options available for customizing your installation. Please follow instruction describe [here](https://tensorflow.rstudio.com/install/).
1. First configure R with a Python installation it can use, like this:
> library(reticulate)
> path_to_python \<- install_python()
> virtualenv_create("r-reticulate", python = path_to_python)
2. Then use install_keras(), which installes Tensorflow, in addition to some commonly used packages like "scipy" and "tensorflow-datasets".
> install.packages("keras")
> library(keras)
> install_keras(envname = "r-reticulate")
3. You can confirm that the installation succeeded with:
> library(tensorflow)
> tf\$constant("Hello Tensorflow!")
#### Load Library
```{r}
#| warning: false
#| error: false
library(tidyverse)
library(tidymodels)
library(tensorflow)
library(keras)
use_virtualenv("r-reticulate")
```
#### Data
In this exercise we will use following synthetic data set and will use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG variables to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
[gp_soil_data_syn.csv](https://www.dropbox.com/s/c63etg7u5qql2y8/gp_soil_data_syn.csv?dl=0)
```{r}
#| warning: false
#| error: false
#| fig.width: 5
#| fig.height: 4
# define file from my github
urlfile = "https://github.com//zia207/r-colab/raw/main/Data/USA/gp_soil_data_syn.csv"
mf<-read_csv(url(urlfile))
# Create a data-frame
df<-mf %>% dplyr::select(SOC, DEM, Slope, TPI,MAT, MAP,NDVI, NLCD, FRG)%>%
glimpse()
```
#### Inspect the data
Review the joint distribution of a few pairs of columns from the training set using GGally package.
```{r}
df_01<-df %>% dplyr::select(SOC, DEM, Slope, TPI, MAT, MAP,NDVI )%>%
glimpse()
```
### Data Pre-processing
```{r}
#| warning: false
#| error: false
dataset <- recipe(SOC ~ ., data = df) %>%
step_zv(all_predictors()) %>%
step_dummy(all_nominal()) %>%
prep() %>%
bake(new_data = NULL) %>%
glimpse()
```
#### Split the data into training and test sets
```{r}
split <- initial_split(dataset, 0.8, strata = SOC)
train_dataset <- training(split)
test_dataset <- testing(split)
```
#### Split features from labels (SOC)
Separate the target or response value---the "label" (SOC) from the features. This label is the value that you will train the model to predict.
```{r}
#| warning: false
#| error: false
train_features <- train_dataset %>% select(-SOC)
test_features <- test_dataset %>% select(-SOC)
train_labels <- train_dataset %>% select(SOC)
test_labels <- test_dataset %>% select(SOC)
```
#### Data Normalization
It is good practice to normalize features that use different scales and ranges.
One reason this is important is because the features are multiplied by the model weights. So, the scale of the outputs and the scale of the gradients are affected by the scale of the inputs.
Although a model *might* converge without feature normalization, normalization makes training much more stable.
Note: There is no advantage to normalizing the one-hot features---it is done here for simplicity. For more details on how to use the preprocessing layers, refer to the [Working with preprocessing layers](https://www.tensorflow.org/guide/keras/preprocessing_layers) guide and the [Classify structured data using Keras preprocessing layers](https://tensorflow.rstudio.com/tutorials/structured_data/preprocessing_layers.qmd) tutorial.
Source:(https://tensorflow.rstudio.com/tutorials/keras/regression#inspect-the-data
#### The Normalization layer
The layer_normalization() is a clean and simple way to add feature normalization into your model.
The first step is to create the layer:
```{r}
#| warning: false
#| error: false
normalizer <- layer_normalization(axis = -1L)
```
Then, fit the state of the preprocessing layer to the data by calling `adapt()`:
```{r}
#| warning: false
#| error: false
normalizer %>% adapt(as.matrix(train_features))
```
#### Duild and Complie Model
- The normalization layer for a multiple-input model).
- Two hidden, non-linear, `Dense` layers with the ReLU (`relu`) activation function nonlinearity.
- A linear `Dense` single-output layer.
Both models will use the same training procedure so the `compile` method is included in the `build_and_compile_model` function below.
```{r}
#| warning: false
#| error: false
build_and_compile_model <- function(norm) {
model <- keras_model_sequential() %>%
norm() %>%
layer_dense(64, activation = 'relu') %>%
layer_dense(64, activation = 'relu') %>%
layer_dense(1)
model %>% compile(
loss = 'mean_absolute_error',
optimizer = optimizer_adam(0.001)
)
model
}
```
```{r}
#| warning: false
#| error: false
dnn_model <- build_and_compile_model(normalizer)
summary(dnn_model)
```
Use Keras fit() to execute the training for 100 epochs:
```{r}
#| warning: false
#| error: false
history <- dnn_model %>% fit(
as.matrix(train_features),
as.matrix(train_labels),
validation_split = 0.1,
verbose = 0,
epochs = 100
)
```
```{r}
#| fig.width: 5
#| fig.height: 4
plot(history)
```
#### Prediction
```{r}
test_results <- list()
test_results[['dnn_model']] <- dnn_model %>% evaluate(
as.matrix(test_features),
as.matrix(test_labels),
verbose = 0
)
```
```{r}
test_results
```
```{r}
#| fig.width: 5
#| fig.height: 4
test_labels$Pred.SOC <- as.data.frame(predict(dnn_model, as.matrix(test_features)))
```
```{r}
names(test_labels)
```
```{r}
RMSE<- Metrics::rmse(test_labels$SOC, test_labels$Pred.SOC$V1)
MAE<- Metrics::mae(test_labels$SOC, test_labels$Pred.SOC$V1)
# Print results
paste0("RMSE: ", round(RMSE,2))
paste0("MAE: ", round(MAE,2))
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 4
library(ggpmisc)
formula<-y~x
ggplot(test_labels, aes(SOC,Pred.SOC$V1)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("DNN-Keras") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))
```
```{r}
# remove all object
rm(list = ls())
```
### Tuning Keras-DNN with Tidymodel
#### Load Library
```{r}
#| warning: false
#| error: false
library(tidyverse)
library(tidymodels)
library(tensorflow)
library(keras)
use_virtualenv("r-reticulate")
```
#### Data
In this exercise we will use following synthetic data set and will use DEM, Slope, TPI, MAT, MAP, NDVI, NLCD, and FRG variables to fit Deep Neural Network regression model. This data was created with AI using gp_soil_data data set
[gp_soil_data_syn.csv](https://www.dropbox.com/s/c63etg7u5qql2y8/gp_soil_data_syn.csv?dl=0)
```{r}
#| warning: false
#| error: false
#| fig.width: 5
#| fig.height: 4
# define file from my github
urlfile = "https://github.com//zia207/r-colab/raw/main/Data/USA/gp_soil_data_syn.csv"
mf<-read_csv(url(urlfile))
# Create a data-frame
df<-mf %>% dplyr::select(SOC, DEM, Slope, TPI,MAT, MAP,NDVI, NLCD, FRG)%>%
glimpse()
```
#### Data split
The data set (n = 1408) will randomly split into sub-sets for training (70%), validation (15%) and test data (15%). The validation data will be used to optimized the model parameters during the tuning and training processes. The test data set will be used as the hold-out data to evaluate the DNN model.
```{r}
#| warning: false
#| error: false
#| fig.width: 4.5
#| fig.height: 4
set.seed(1245) # for reproducibility
library(tidymodels)
split <- initial_split(df, prop = 0.8, strata = SOC)
train <- split %>% training()
test <- split %>% testing()
```
#### Data Pre-processing
```{r}
dnn_recipe <-
recipe(SOC ~ ., data = train) %>%
step_zv(all_predictors()) %>%
step_dummy(all_nominal()) %>%
step_normalize(all_numeric_predictors())
# For testing when we arrive at a final model:
test_normalized <- bake(prep(dnn_recipe), new_data = test, all_predictors())
# Set 10 fold cross-validation data set
cv_folds<- vfold_cv(train, v=5)
```
#### Specify tunable Hypermeters of Keras - DNN
```{r}
#| warning: false
#| error: false
dnn_model <-
mlp(mode = "regression",
hidden_units = tune(),
dropout = tune(),
epochs = tune(),
activation = "relu",
) %>%
set_engine("keras",
verbose = 0,
optimizer = optimizer_adam(0.001),
validation = .10)
dnn_model
```
#### Create Workflow
```{r}
#| warning: false
#| error: false
dnn_wf <- workflow() %>%
add_model(dnn_model) %>%
add_recipe(dnn_recipe)
dnn_wf
```
#### Define random grid prameters
```{r}
#| warning: false
#| error: false
dnn_grid <- parameters(dnn_model) %>%
finalize(train) %>%
grid_random(size = 10)
head(dnn_grid)
```
#### Tune Grid
```{r}
#| warning: false
#| error: false
#doParallel::registerDoParallel()
set.seed(345)
# grid search
dnn_tune_grid <- dnn_wf %>%
tune_grid(
resamples = cv_folds,
grid = dnn_grid,
control = control_grid(verbose = F),
metrics = metric_set(rmse, rsq, mae)
)
```
```{r}
#| warning: false
#| error: false
collect_metrics(dnn_tune_grid )
```
```{r}
#| warning: false
#| error: false
#| fig.width: 8
#| fig.height: 7
autoplot(dnn_tune_grid)
```
#### The best DNN model
```{r}
#| warning: false
#| error: false
best_rmse <- select_best(dnn_tune_grid , "rmse")
dnn_final <- finalize_model(
dnn_model,
best_rmse
)
dnn_final
```
#### Fit the model
```{r}
#| warning: false
#| error: false
final_fit <- fit(dnn_final, SOC ~ ., data = train)
```
#### Prediction
```{r}
test$SOC.pred = predict(final_fit, test)
```
```{r}
#| warning: false
#| error: false
library(Metrics)
RMSE<- Metrics::rmse(test$SOC, test$SOC.pred$.pred)
RMSE
```
```{r}
#| warning: false
#| error: false
#| fig.width: 4
#| fig.height: 4
library(ggpmisc)
formula<-y~x
ggplot(test, aes(SOC,SOC.pred$.pred)) +
geom_point() +
geom_smooth(method = "lm")+
stat_poly_eq(use_label(c("eq", "adj.R2")), formula = formula) +
ggtitle("DNN-Keras") +
xlab("Observed") + ylab("Predicted") +
scale_x_continuous(limits=c(0,25), breaks=seq(0, 25, 5))+
scale_y_continuous(limits=c(0,25), breaks=seq(0, 25, 5)) +
# Flip the bars
theme(
panel.background = element_rect(fill = "grey95",colour = "gray75",size = 0.5, linetype = "solid"),
axis.line = element_line(colour = "grey"),
plot.title = element_text(size = 14, hjust = 0.5),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
axis.text.x=element_text(size=13, colour="black"),
axis.text.y=element_text(size=13,angle = 90,vjust = 0.5, hjust=0.5, colour='black'))
```
### Further Reading
1. [Quick start](https://tensorflow.rstudio.com/install/)
2. [Basic Regression with Keras-Tensorflow](https://tensorflow.rstudio.com/tutorials/keras/regression#inspect-the-data)
3. [Multilayer perceptron via keras](https://parsnip.tidymodels.org/reference/details_mlp_keras.html)