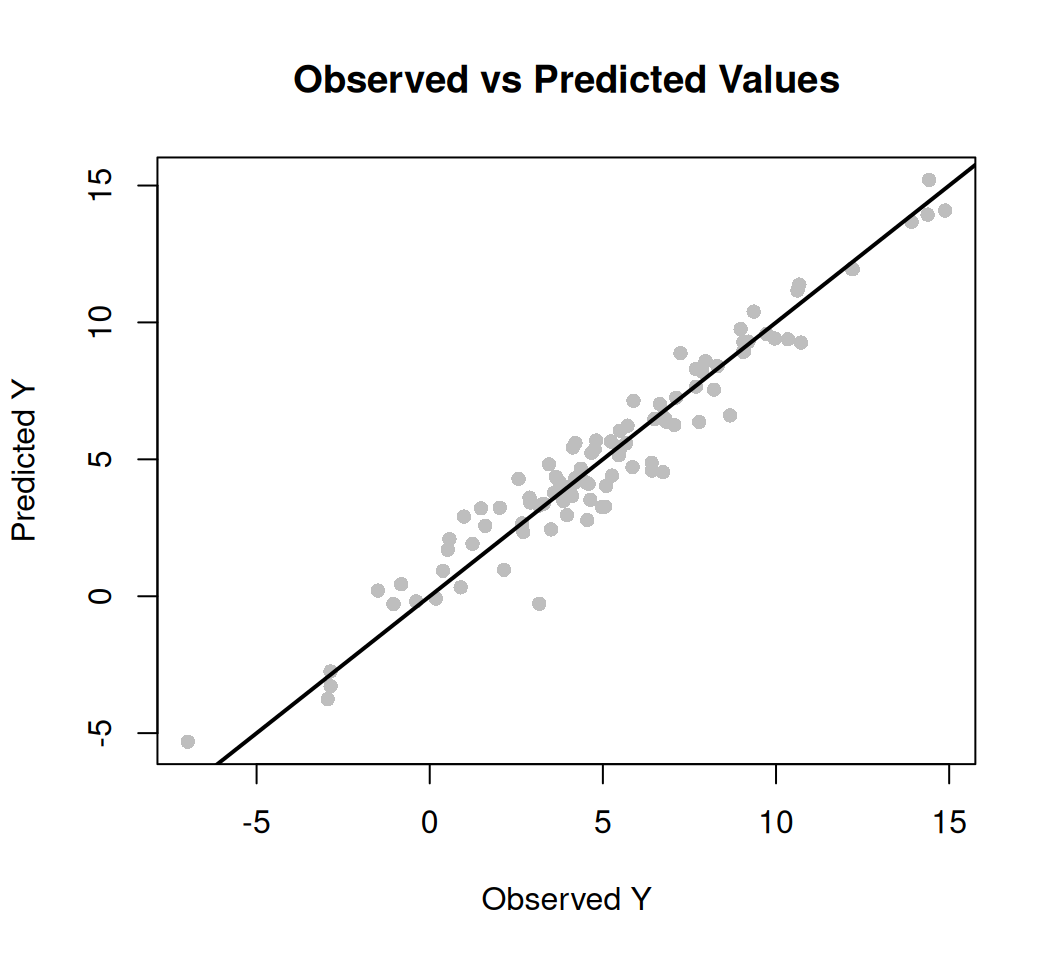
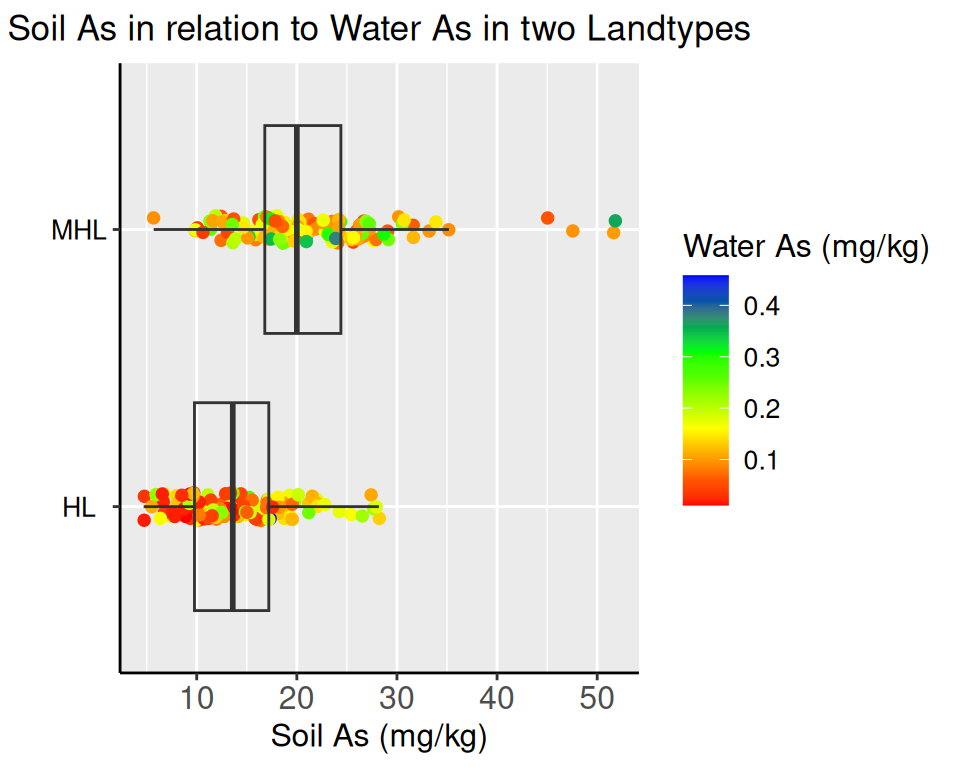
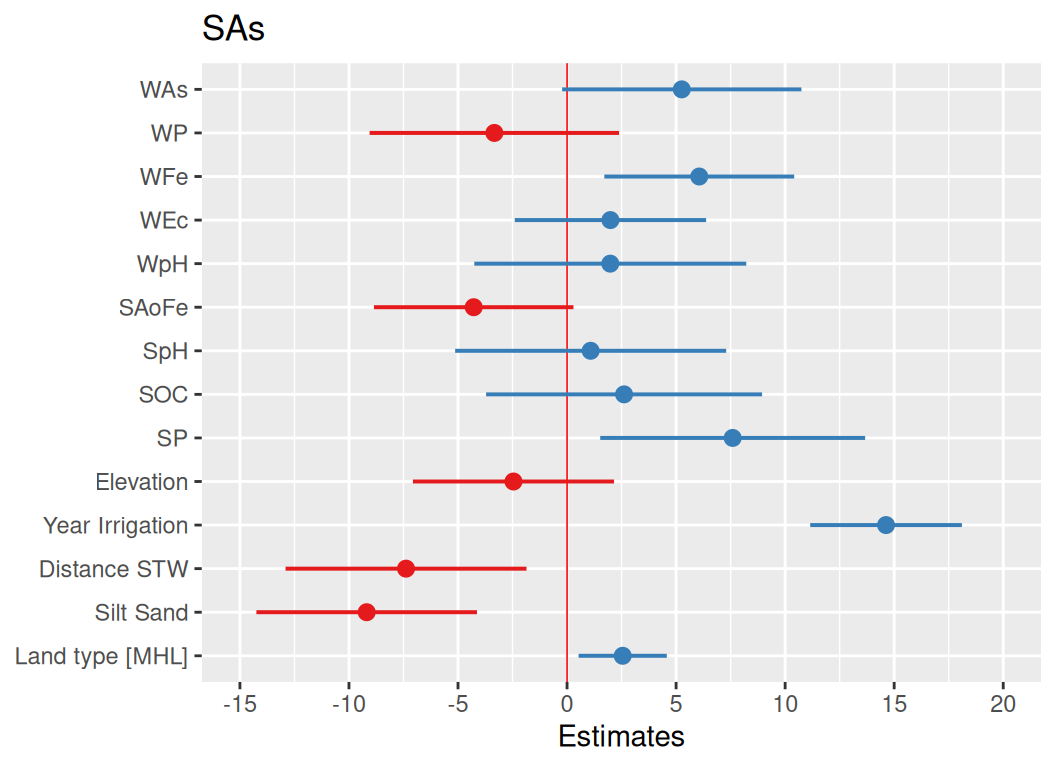
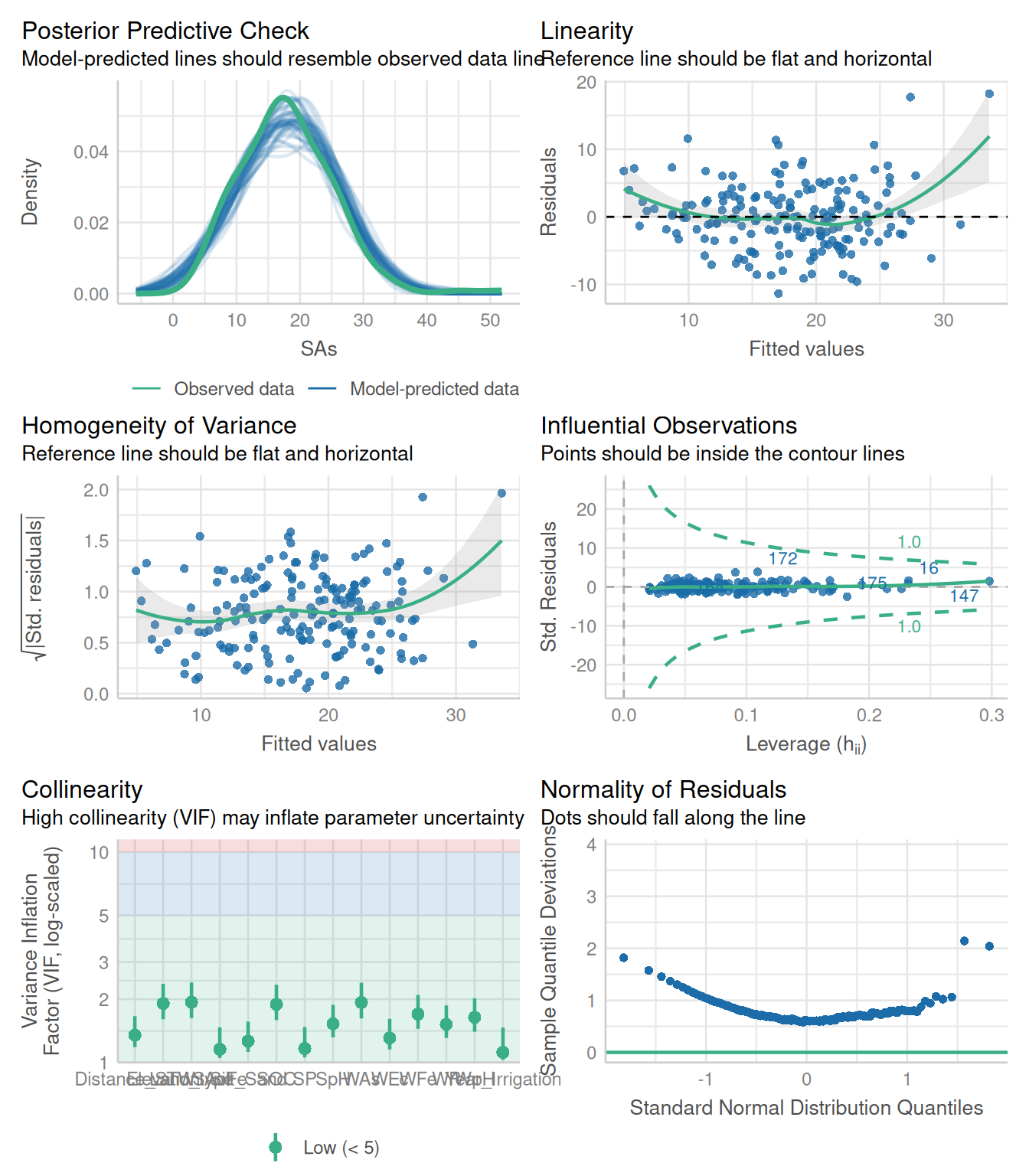
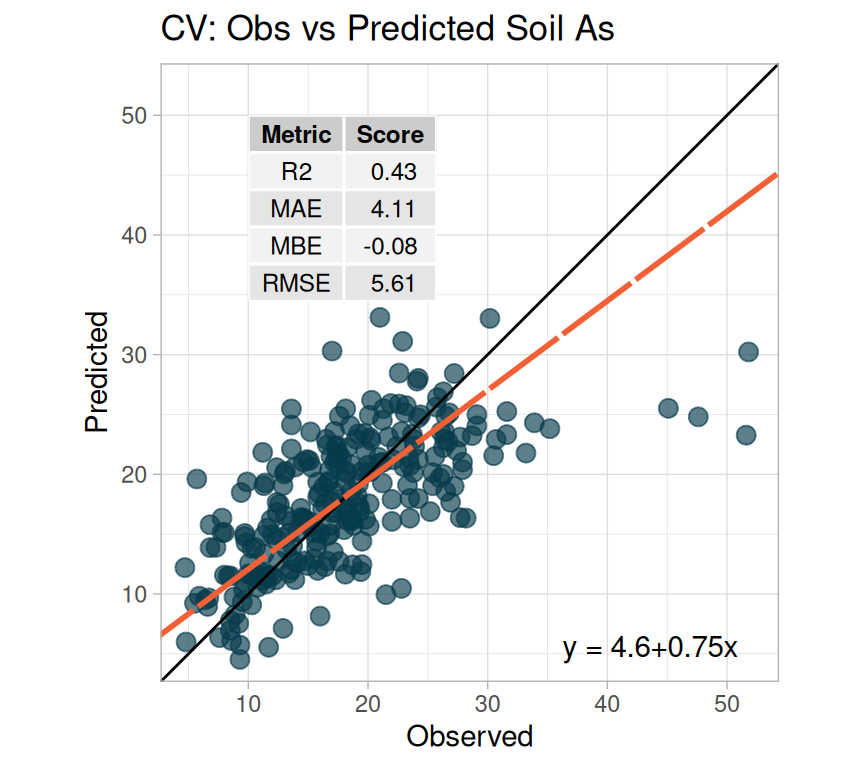
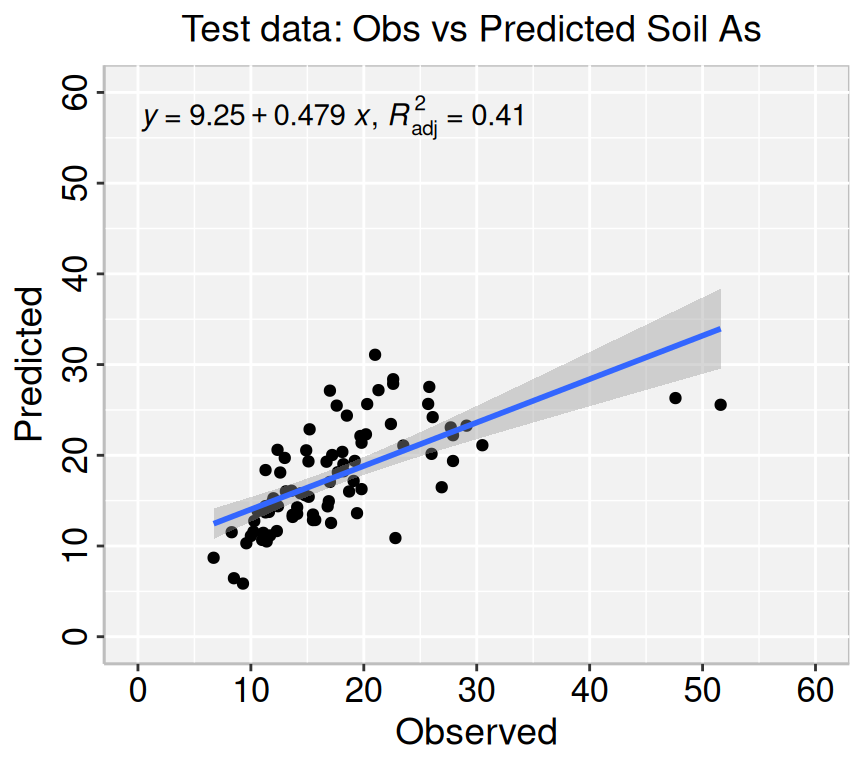
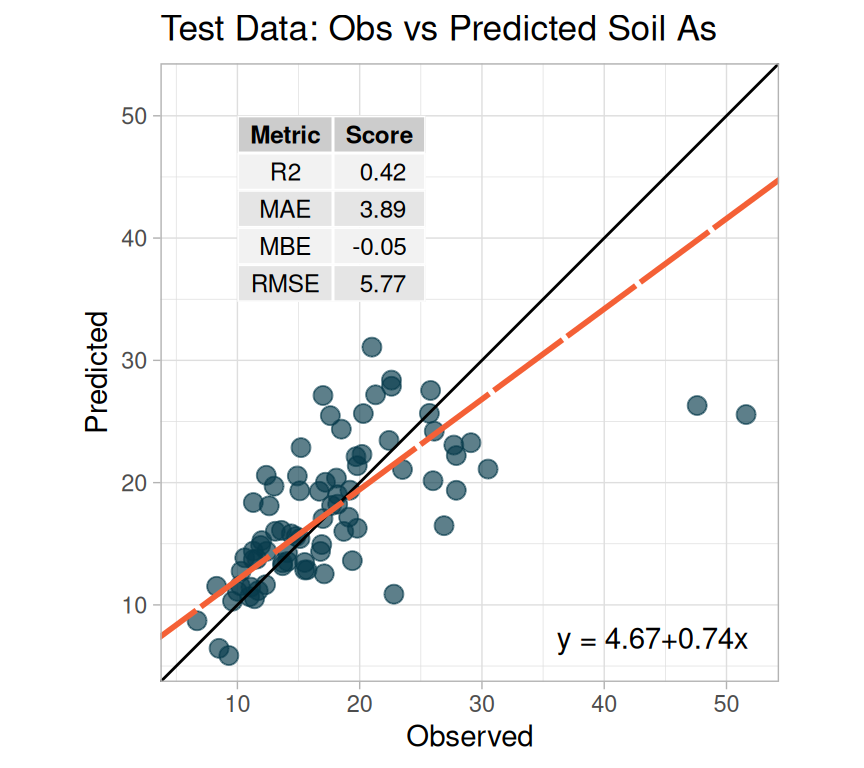
We fitted a linear model (estimated using ML) to predict SAs with WAs, WP, WFe,
WEc, WpH, SAoFe, SpH, SOC, SP, Elevation, Year_Irrigation, Distance_STW,
Silt_Sand and Land_type (formula: SAs ~ WAs + WP + WFe + WEc + WpH + SAoFe +
SpH + SOC + SP + Elevation + Year_Irrigation + Distance_STW + Silt_Sand +
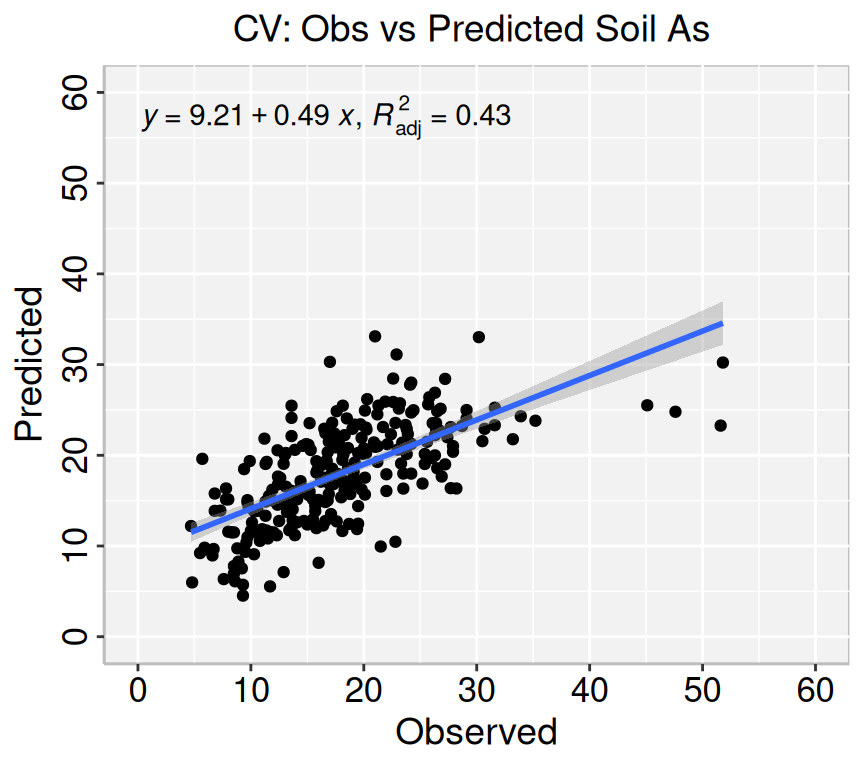
Land_type). The model's explanatory power is substantial (R2 = 0.57). The
model's intercept, corresponding to WAs = 0, WP = 0, WFe = 0, WEc = 0, WpH = 0,
SAoFe = 0, SpH = 0, SOC = 0, SP = 0, Elevation = 0, Year_Irrigation = 0,
Distance_STW = 0, Silt_Sand = 0 and Land_type = HL, is at 13.04 (95% CI [4.64,
21.45], t(168) = 3.04, p = 0.002). Within this model:
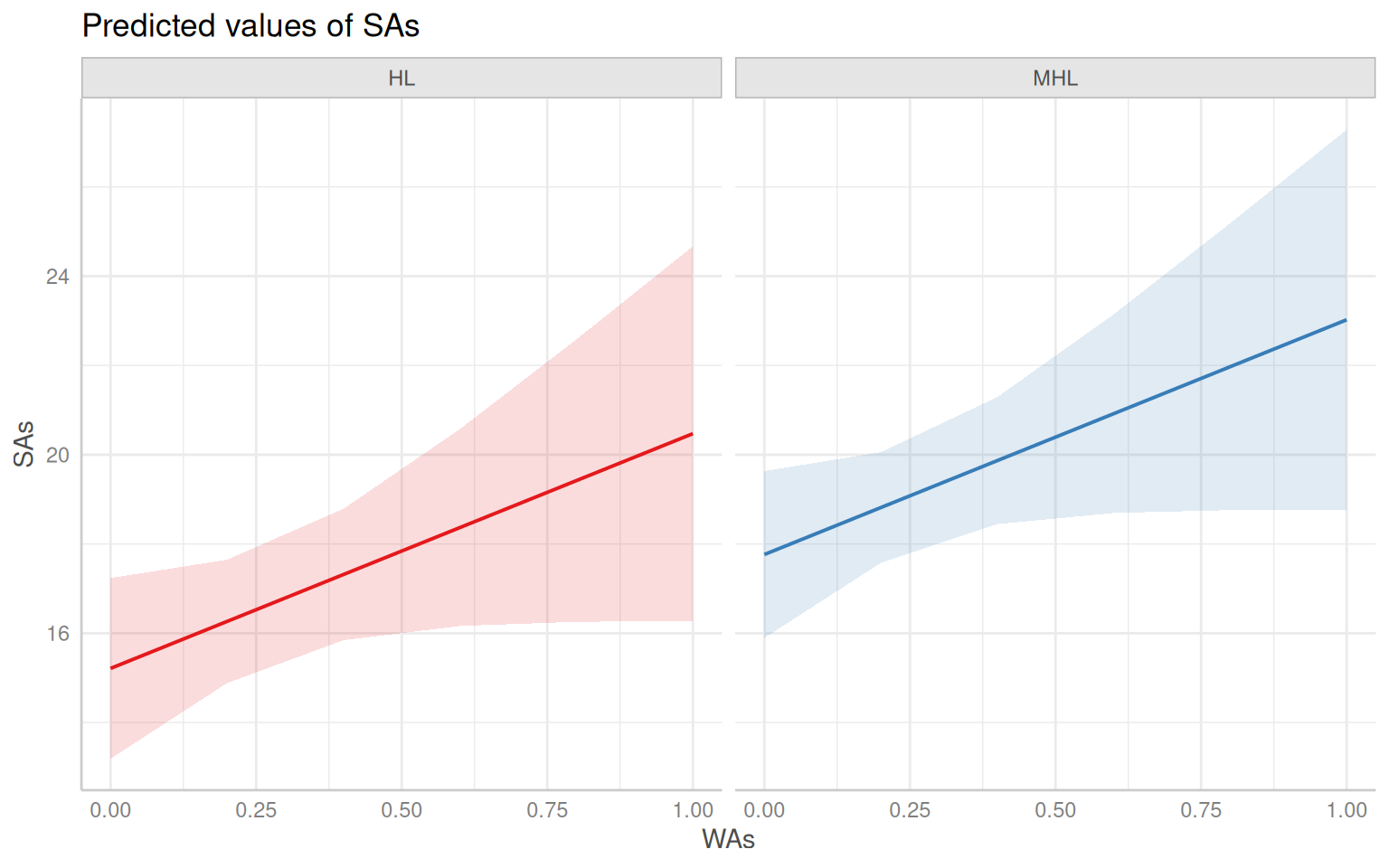
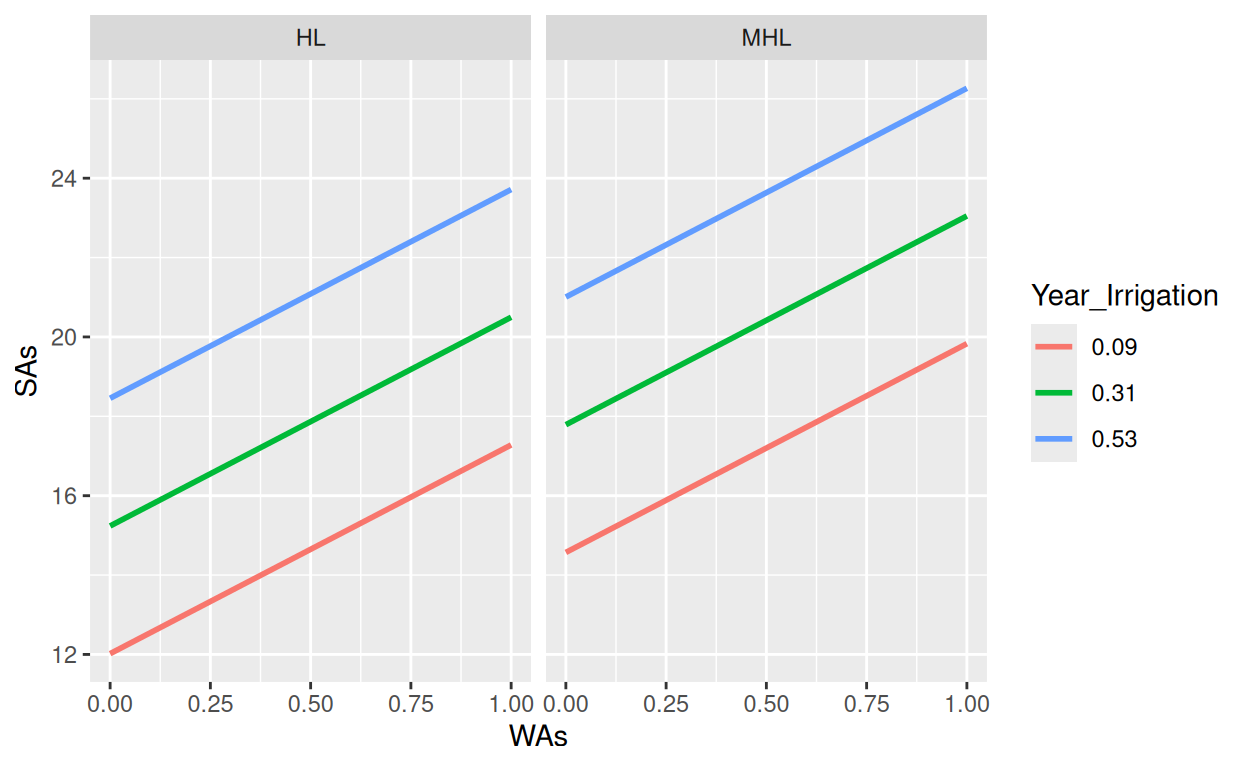
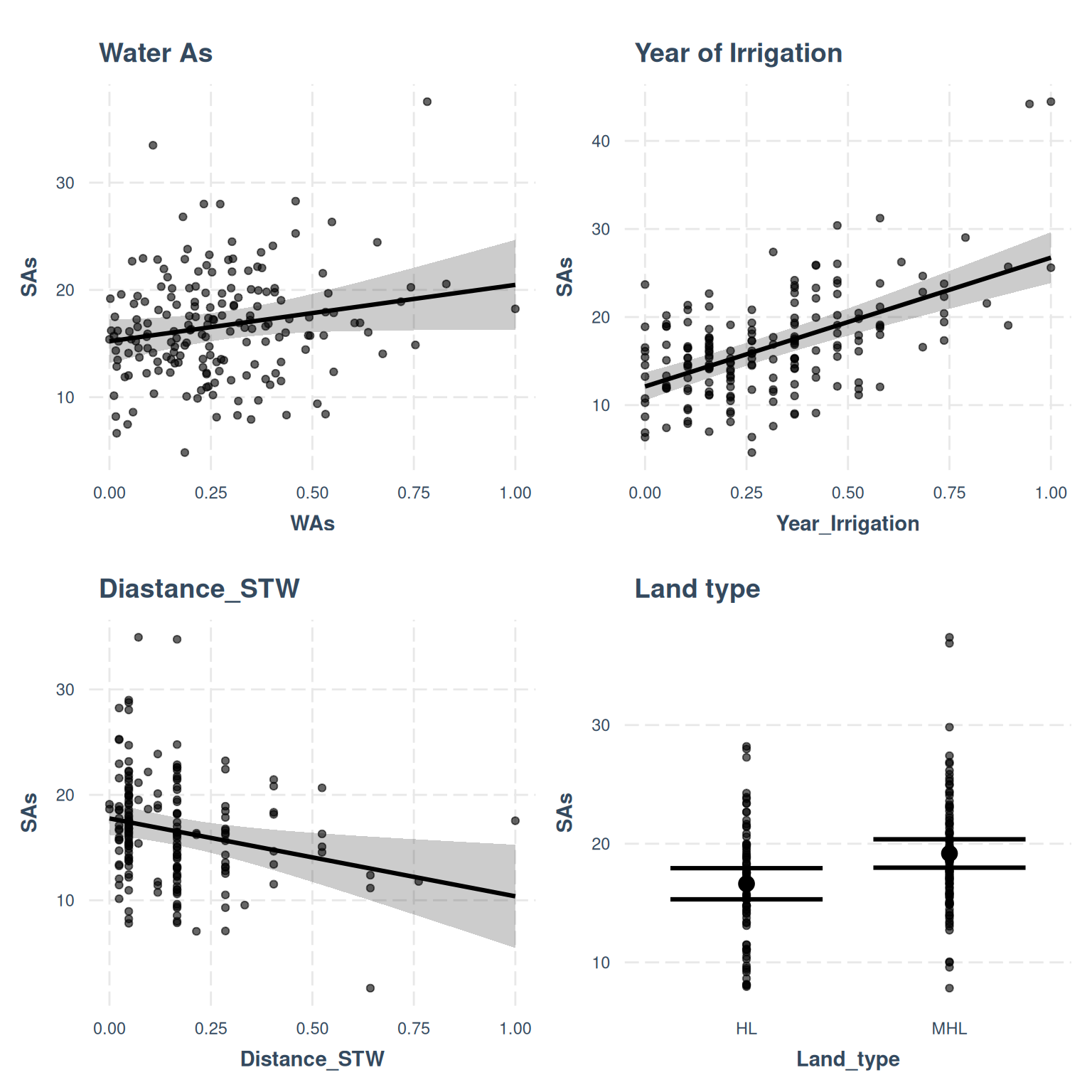
- The effect of WAs is statistically non-significant and positive (beta = 5.26,
95% CI [-0.23, 10.75], t(168) = 1.88, p = 0.060; Std. beta = 0.13, 95% CI
[-5.70e-03, 0.27])
- The effect of WP is statistically non-significant and negative (beta = -3.33,
95% CI [-9.05, 2.39], t(168) = -1.14, p = 0.253; Std. beta = -0.07, 95% CI
[-0.19, 0.05])
- The effect of WFe is statistically significant and positive (beta = 6.06, 95%
CI [1.71, 10.42], t(168) = 2.73, p = 0.006; Std. beta = 0.18, 95% CI [0.05,
0.31])
- The effect of WEc is statistically non-significant and positive (beta = 1.99,
95% CI [-2.40, 6.38], t(168) = 0.89, p = 0.374; Std. beta = 0.05, 95% CI
[-0.06, 0.16])
- The effect of WpH is statistically non-significant and positive (beta = 1.98,
95% CI [-4.25, 8.22], t(168) = 0.62, p = 0.533; Std. beta = 0.04, 95% CI
[-0.09, 0.17])
- The effect of SAoFe is statistically non-significant and negative (beta =
-4.28, 95% CI [-8.85, 0.29], t(168) = -1.83, p = 0.067; Std. beta = -0.10, 95%
CI [-0.21, 6.85e-03])
- The effect of SpH is statistically non-significant and positive (beta = 1.08,
95% CI [-5.13, 7.30], t(168) = 0.34, p = 0.733; Std. beta = 0.02, 95% CI
[-0.10, 0.14])
- The effect of SOC is statistically non-significant and positive (beta = 2.61,
95% CI [-3.71, 8.94], t(168) = 0.81, p = 0.418; Std. beta = 0.06, 95% CI
[-0.08, 0.19])
- The effect of SP is statistically significant and positive (beta = 7.59, 95%
CI [1.52, 13.67], t(168) = 2.45, p = 0.014; Std. beta = 0.13, 95% CI [0.03,
0.24])
- The effect of Elevation is statistically non-significant and negative (beta =
-2.46, 95% CI [-7.07, 2.15], t(168) = -1.04, p = 0.296; Std. beta = -0.07, 95%
CI [-0.21, 0.06])
- The effect of Year Irrigation is statistically significant and positive (beta
= 14.63, 95% CI [11.15, 18.10], t(168) = 8.25, p < .001; Std. beta = 0.44, 95%
CI [0.34, 0.55])
- The effect of Distance STW is statistically significant and negative (beta =
-7.38, 95% CI [-12.91, -1.86], t(168) = -2.62, p = 0.009; Std. beta = -0.15,
95% CI [-0.27, -0.04])
- The effect of Silt Sand is statistically significant and negative (beta =
-9.19, 95% CI [-14.24, -4.13], t(168) = -3.56, p < .001; Std. beta = -0.20, 95%
CI [-0.31, -0.09])
- The effect of Land type [MHL] is statistically significant and positive (beta
= 2.55, 95% CI [0.53, 4.57], t(168) = 2.47, p = 0.013; Std. beta = 0.35, 95% CI
[0.07, 0.62])
Standardized parameters were obtained by fitting the model on a standardized
version of the dataset. 95% Confidence Intervals (CIs) and p-values were
computed using a Wald t-distribution approximation.