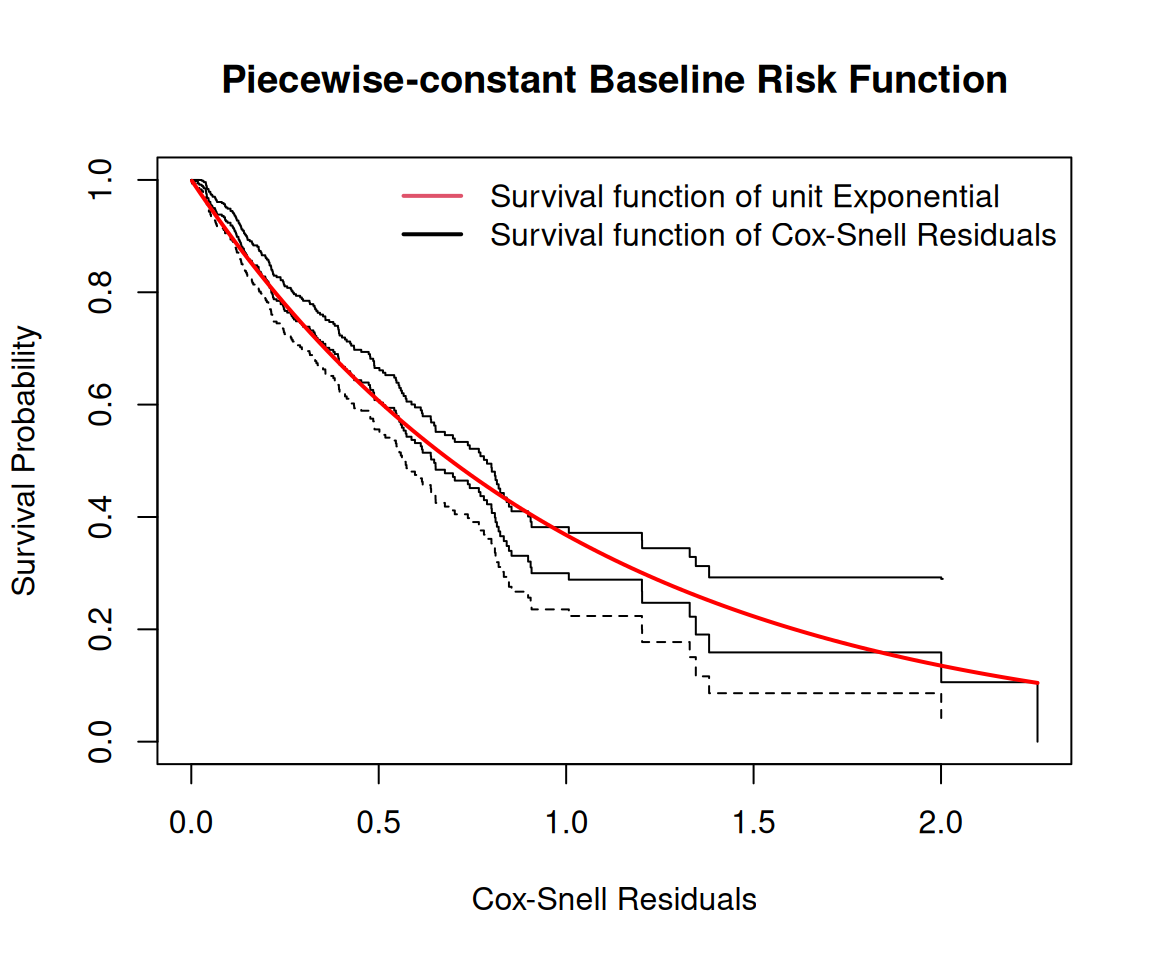
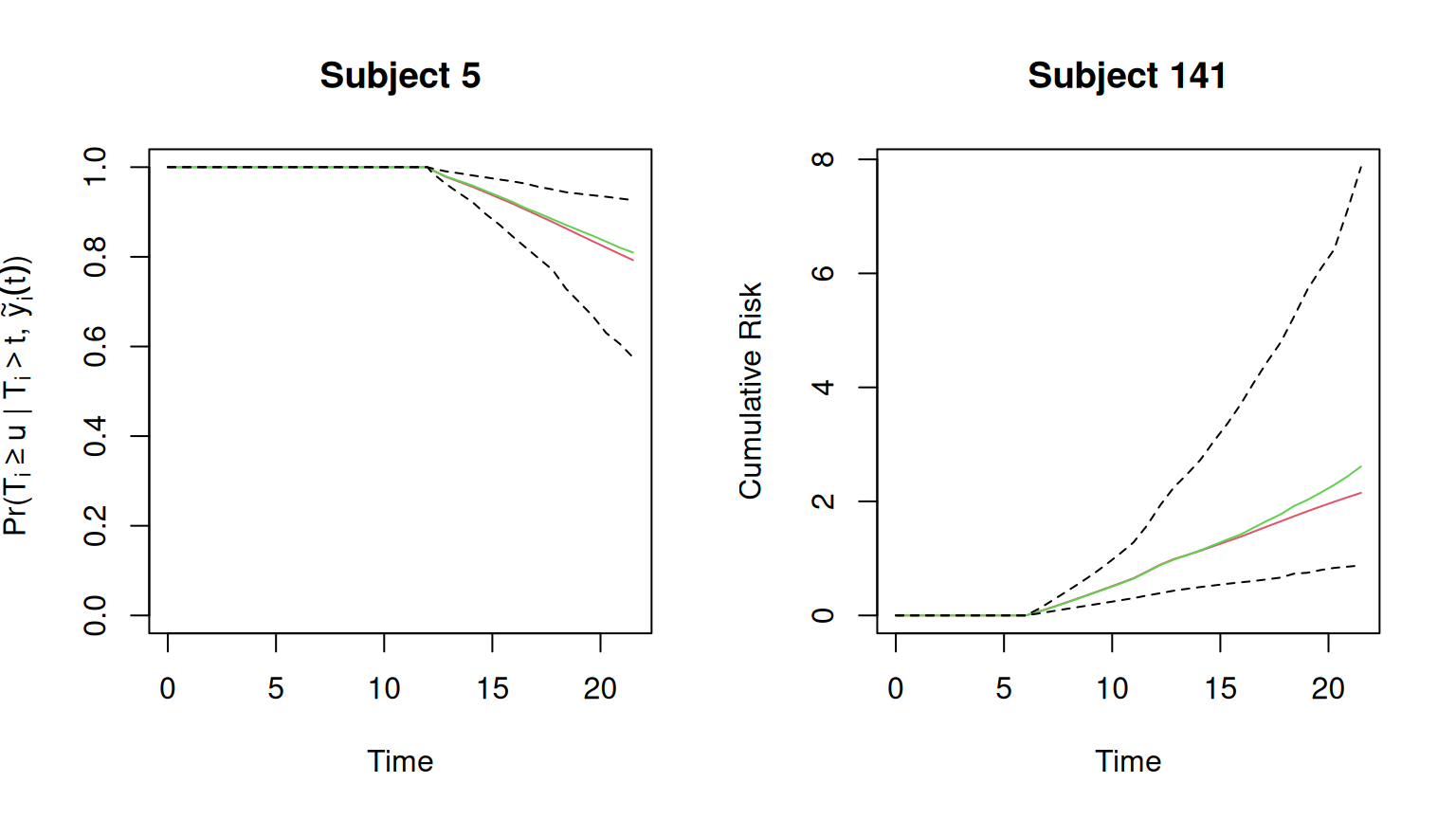
Rows: 467
Columns: 12
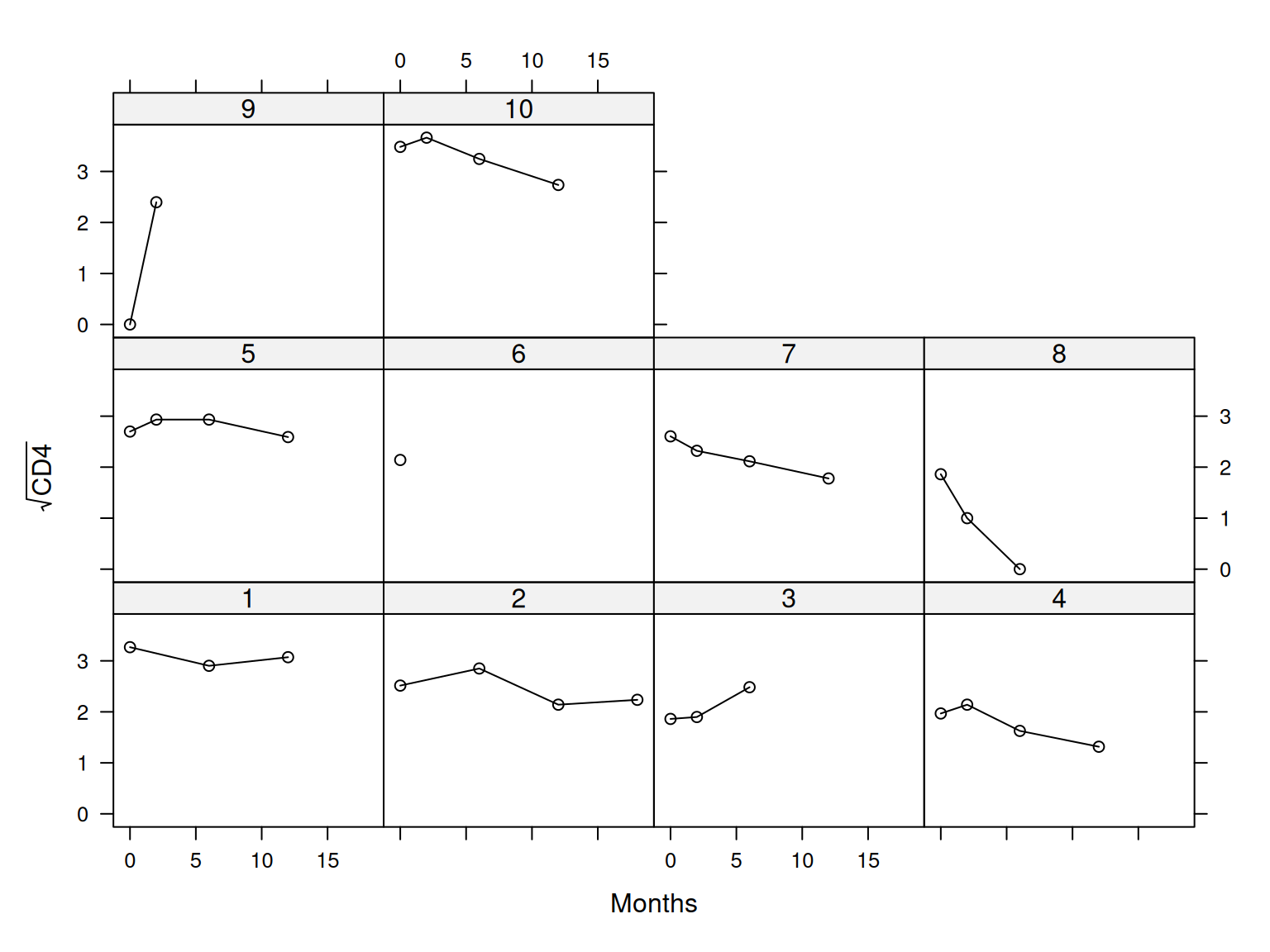
$ patient <fct> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,…
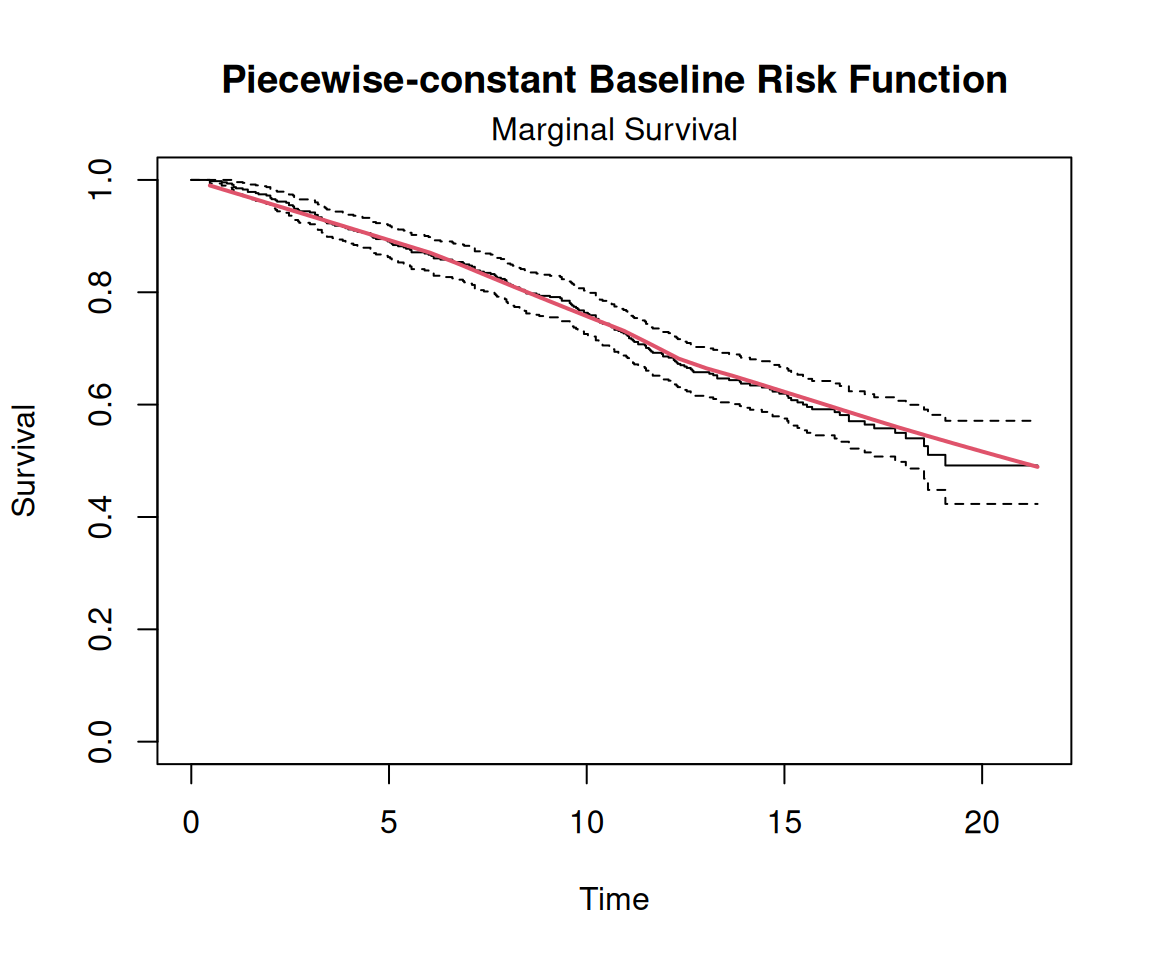
$ Time <dbl> 16.97, 19.00, 18.53, 12.70, 15.13, 1.90, 14.33, 9.57, 11.57, 1…
$ death <int> 0, 0, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0, 1,…
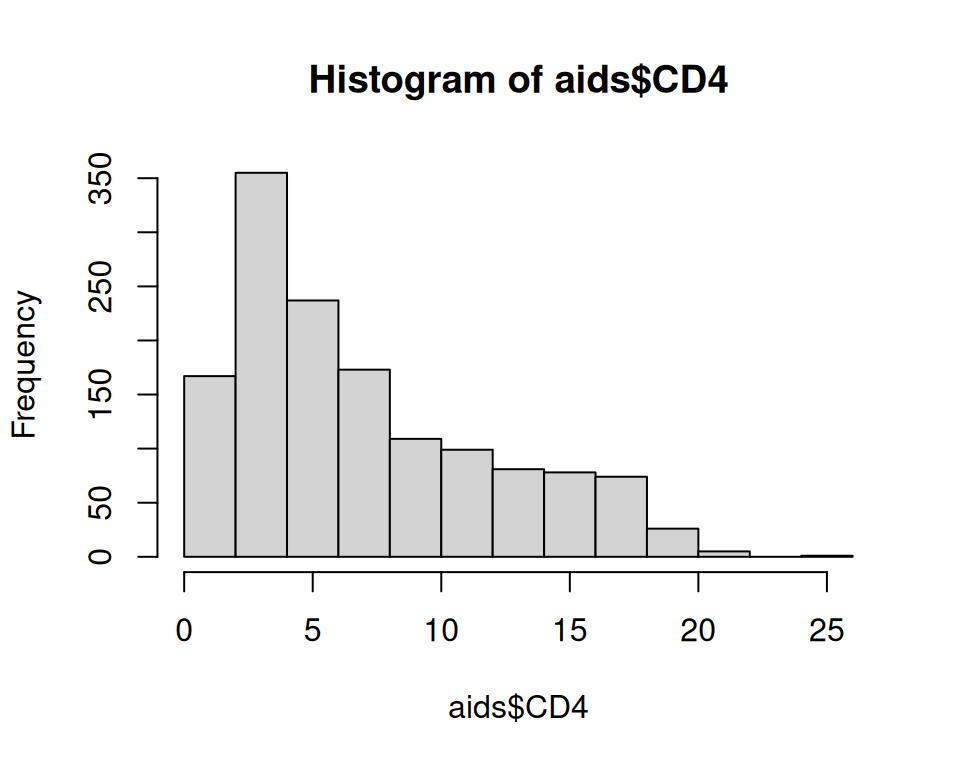
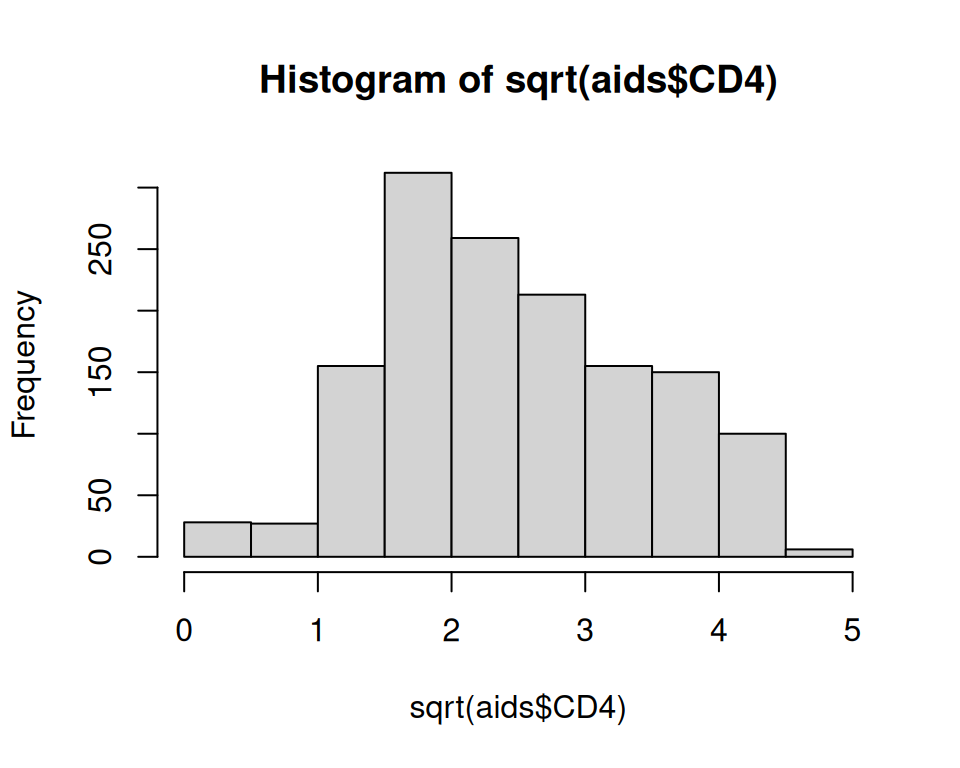
$ CD4 <dbl> 10.677078, 6.324555, 3.464102, 3.872983, 7.280110, 4.582576, 6…
$ obstime <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ drug <fct> ddC, ddI, ddI, ddC, ddI, ddC, ddC, ddI, ddC, ddI, ddC, ddI, dd…
$ gender <fct> male, male, female, male, male, female, male, female, male, ma…
$ prevOI <fct> AIDS, noAIDS, AIDS, AIDS, AIDS, AIDS, AIDS, noAIDS, AIDS, AIDS…
$ AZT <fct> intolerance, intolerance, intolerance, failure, failure, failu…
$ start <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ stop <dbl> 6.00, 6.00, 2.00, 2.00, 2.00, 1.90, 2.00, 2.00, 2.00, 2.00, 2.…
$ event <dbl> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0,…