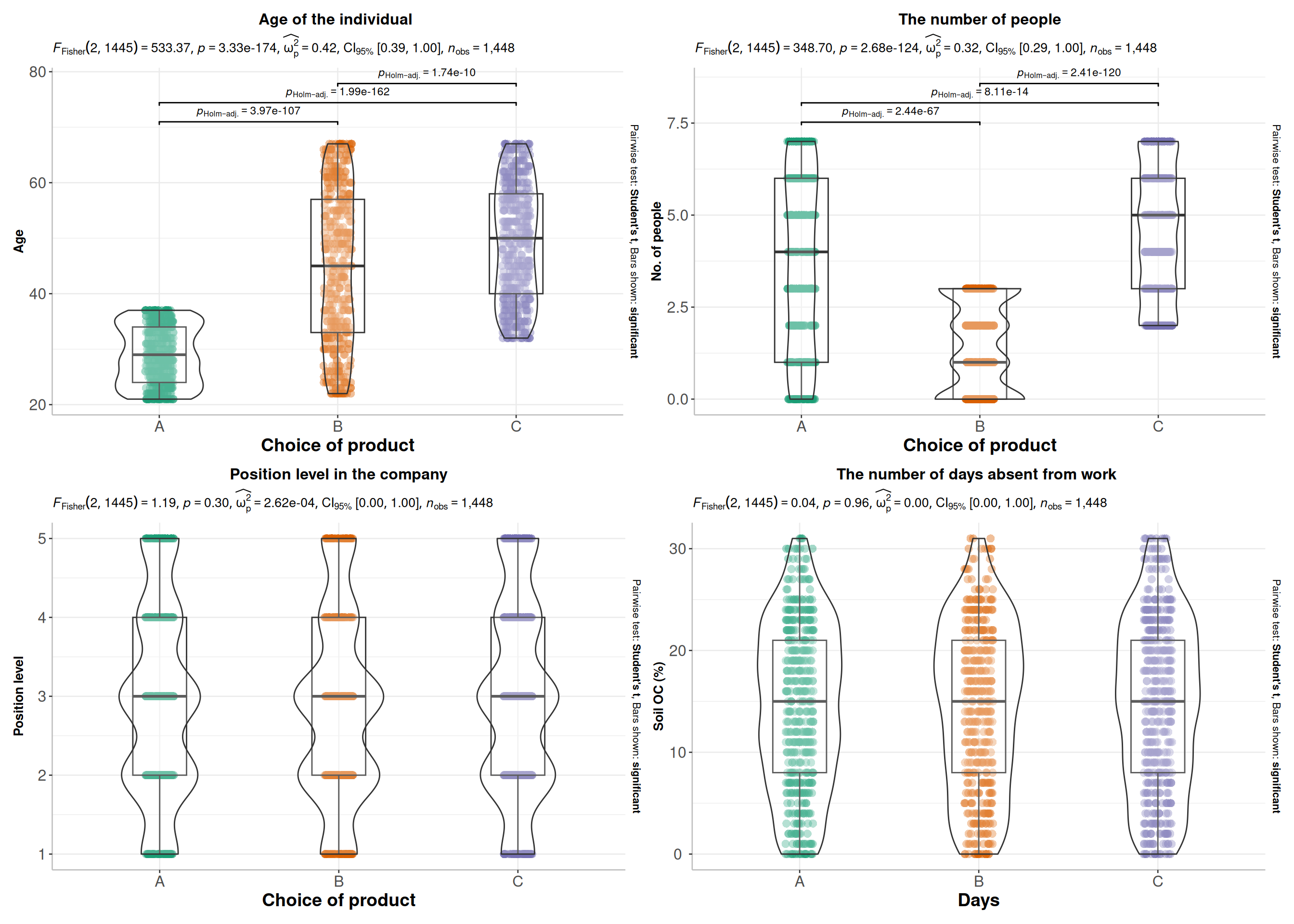
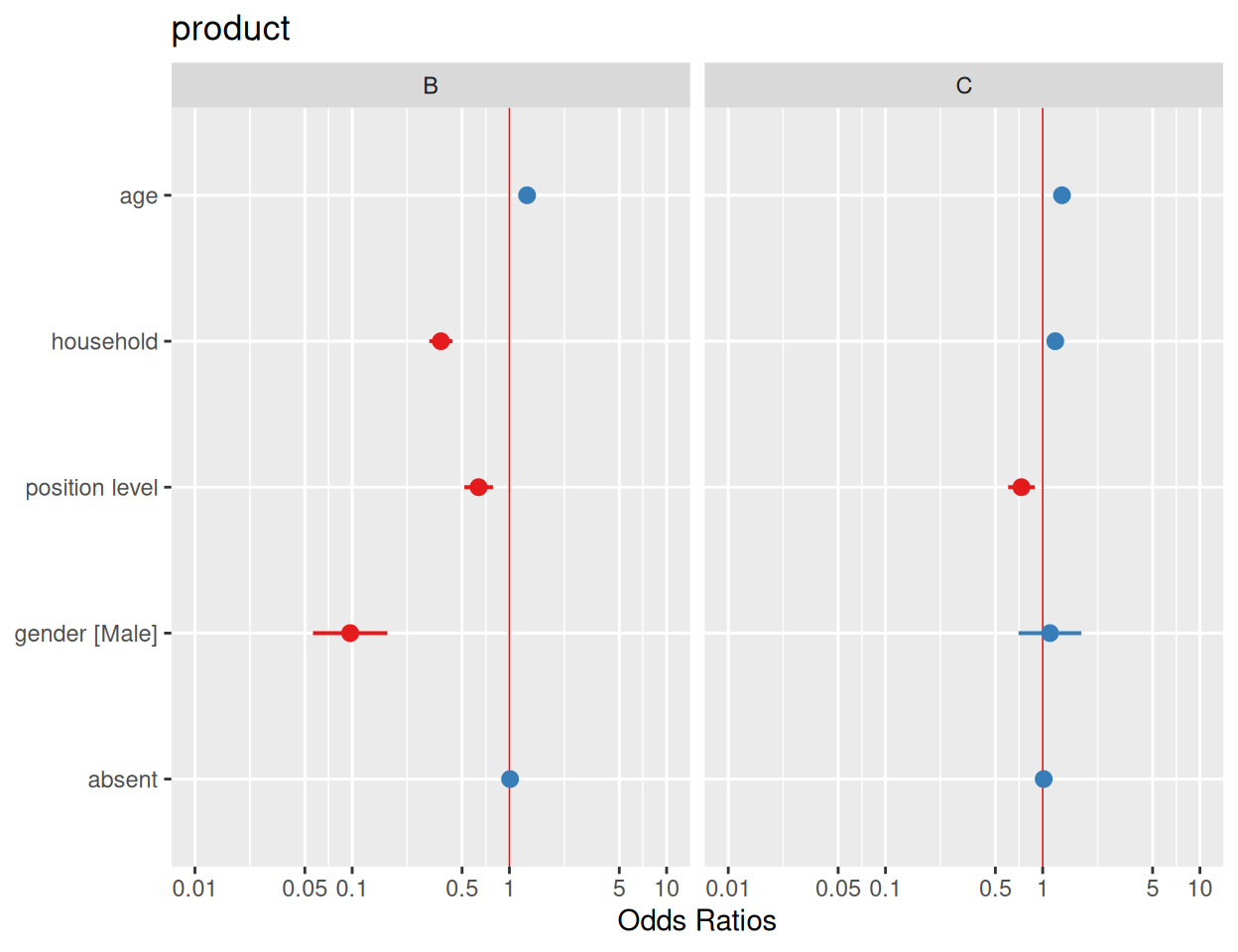
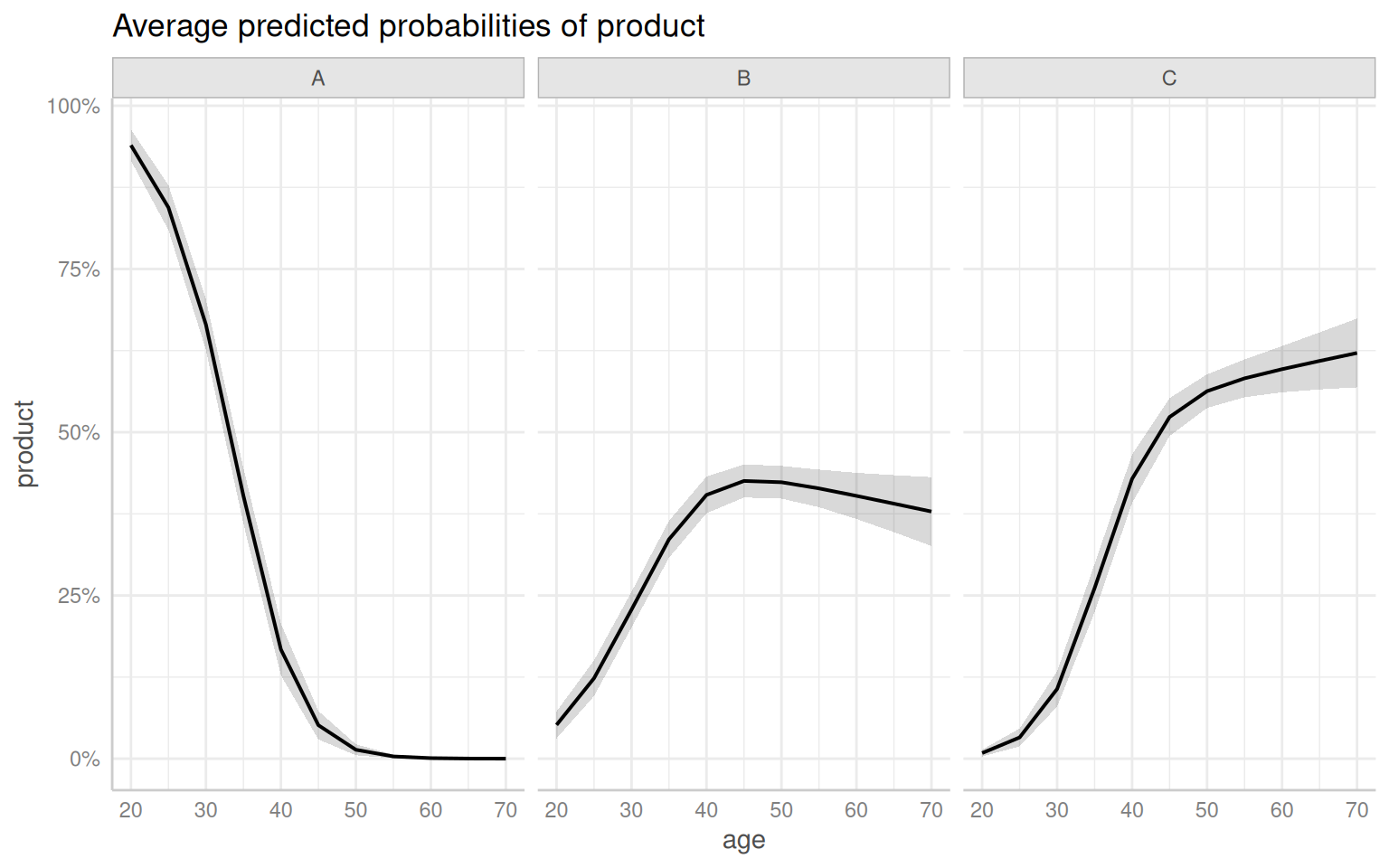
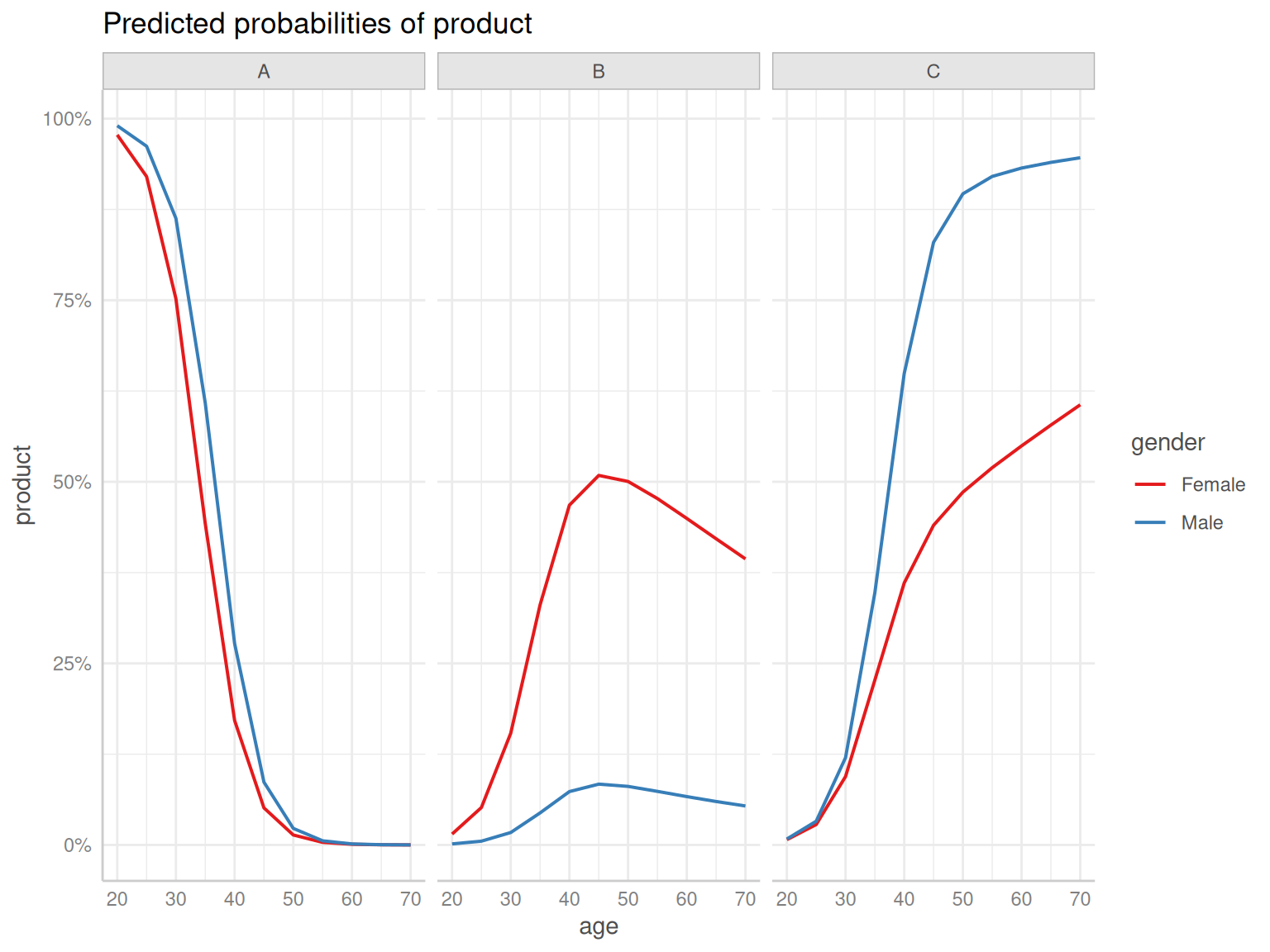
Code
set.seed(123) # For reproducibility
# Create synthetic dataset
n <- 300 # Number of observations
data <- data.frame(
response = factor(sample(c("1", "2", "3"), n, replace = TRUE)),
continuous1 = rnorm(n),
continuous2 = rnorm(n),
continuous3 = rnorm(n),
continuous4 = rnorm(n),
gender = factor(sample(c("Male", "Female"), n, replace = TRUE))
)
# Convert response to a factor and set reference level
data$response <- relevel(data$response, ref = "1")
head(data) response continuous1 continuous2 continuous3 continuous4 gender
1 3 1.9009001 -0.2043353 -0.00497512 0.81293718 Male
2 3 0.7089544 0.5583362 -1.38088679 0.41497661 Female
3 3 0.7361948 0.3242106 -1.25652688 -0.08585906 Male
4 2 1.3657766 0.6977370 0.32524572 -1.90820891 Male
5 3 -0.5762639 -1.0225731 -0.27700053 0.66325198 Male
6 2 -0.8047323 -0.1682708 -0.38901100 0.66168573 Male