Code
# Packages List
packages <- c(
"tidyverse", # Includes readr, dplyr, ggplot2, etc.
"nlme" # For non-linear mixed-effects models
)SSlogisSSfplSSweibullSSgompertzSSasympSSmicmenSelf-starting functions in R are special functions used in non-linear regression models that make the fitting process easier. These functions provide initial parameter estimates that help the fitting algorithm converge more efficiently. Non-linear models often require good starting values for the parameters, and self-starting functions automate this process.
Self-starting functions in R automate the estimation of initial parameters for non-linear models, eliminating the need for manual specification. They are particularly useful in functions like nls() (non-linear least squares) where convergence heavily depends on starting values. Below are key self-starting functions, their descriptions, examples, and visualizations.
#| warning: false
#| error: false
# Install missing packages
new_packages <- packages[!(packages %in% installed.packages()[,"Package"])]
if(length(new_packages)) install.packages(new_packages)
# Verify installation
cat("Installed packages:\n")
print(sapply(packages, requireNamespace, quietly = TRUE))Successfully loaded packages: [1] "package:nlme" "package:lubridate" "package:forcats"
[4] "package:stringr" "package:dplyr" "package:purrr"
[7] "package:readr" "package:tidyr" "package:tibble"
[10] "package:ggplot2" "package:tidyverse" "package:stats"
[13] "package:graphics" "package:grDevices" "package:utils"
[16] "package:datasets" "package:methods" "package:base" SSlogis$y = $
Asym: Asymptote (maximum value).xmid: x-value at the inflection point.scal: Scale parameter (inverse of the growth rate).
Formula: y ~ SSlogis(x, Asym, xmid, scal)
Parameters:
Estimate Std. Error t value Pr(>|t|)
Asym 9.9754 0.4016 24.842 8.43e-15 ***
xmid 9.8424 0.4084 24.098 1.39e-14 ***
scal 3.0210 0.3059 9.875 1.86e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.5136 on 17 degrees of freedom
Number of iterations to convergence: 0
Achieved convergence tolerance: 1.267e-06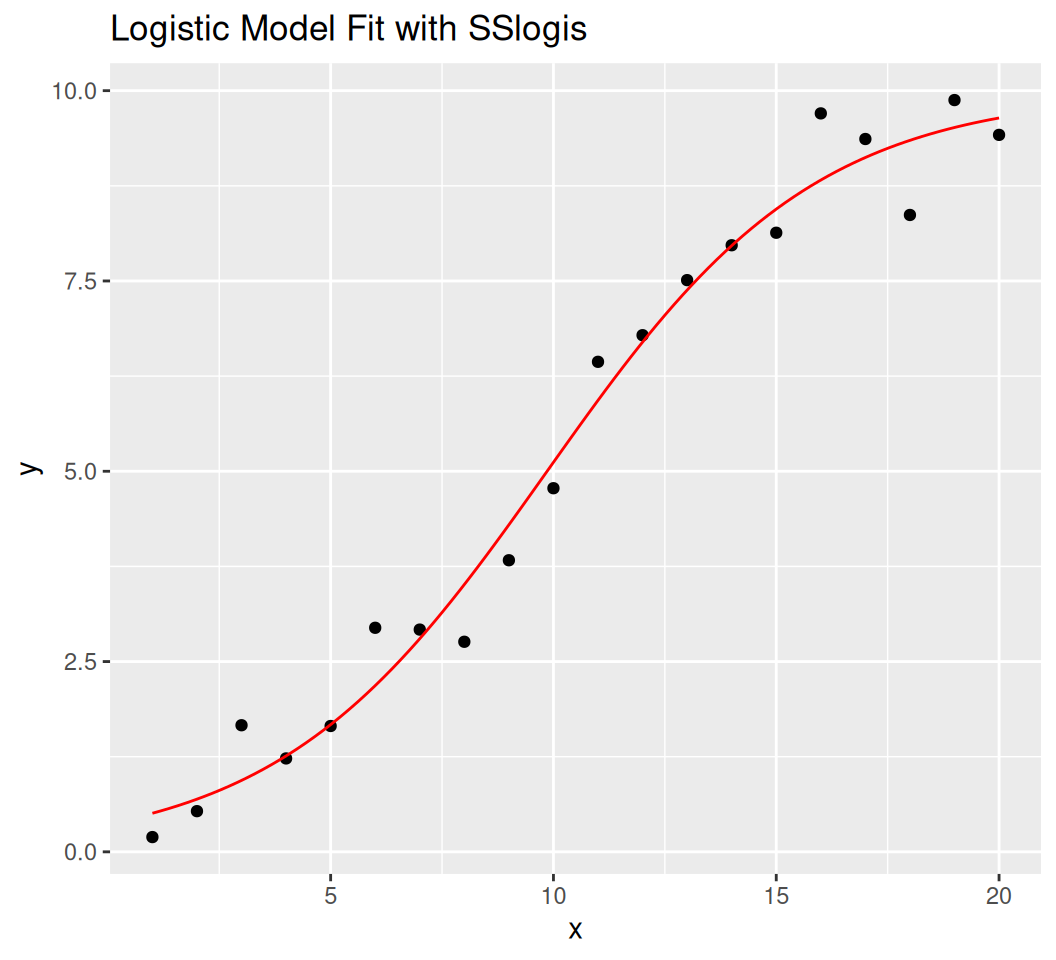
SSfplModel:
\(y = A + \frac{B - A}{1 + (x/xmid)^scal}\)
Parameters:
A: Lower asymptote.B: Upper asymptote.xmid: x-value at the inflection point.scal: Slope parameter.
Formula: y ~ SSfpl(x, A, B, xmid, scal)
Parameters:
Estimate Std. Error t value Pr(>|t|)
A 2.27336 0.14283 15.92 < 2e-16 ***
B 9.94942 0.15096 65.91 < 2e-16 ***
xmid 5.12581 0.08473 60.50 < 2e-16 ***
scal 0.84530 0.07820 10.81 3.24e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4551 on 46 degrees of freedom
Number of iterations to convergence: 0
Achieved convergence tolerance: 1.287e-06# Predict values for a smooth curve
new_x <- data.frame(x = seq(0, 10, length.out = 100))
pred_y <- predict(fit, newdata = new_x)
# Plot
ggplot(df, aes(x, y)) +
geom_point(size = 2, alpha = 0.7) +
geom_line(data = data.frame(x = new_x$x, y = pred_y),
color = "red", linewidth = 1) +
labs(
title = "Four-Parameter Logistic Model (SSfpl)",
x = "x",
y = "Response"
) +
geom_hline(yintercept = coef(fit)["A"], linetype = "dashed", color = "blue") +
geom_hline(yintercept = coef(fit)["B"], linetype = "dashed", color = "blue") +
annotate("text", x = 1, y = coef(fit)["A"] + 0.5,
label = paste0("Lower Asym (A) = ", round(coef(fit)["A"], 2)), color = "blue") +
annotate("text", x = 1, y = coef(fit)["B"] - 0.5,
label = paste0("Upper Asym (B) = ", round(coef(fit)["B"], 2)), color = "blue")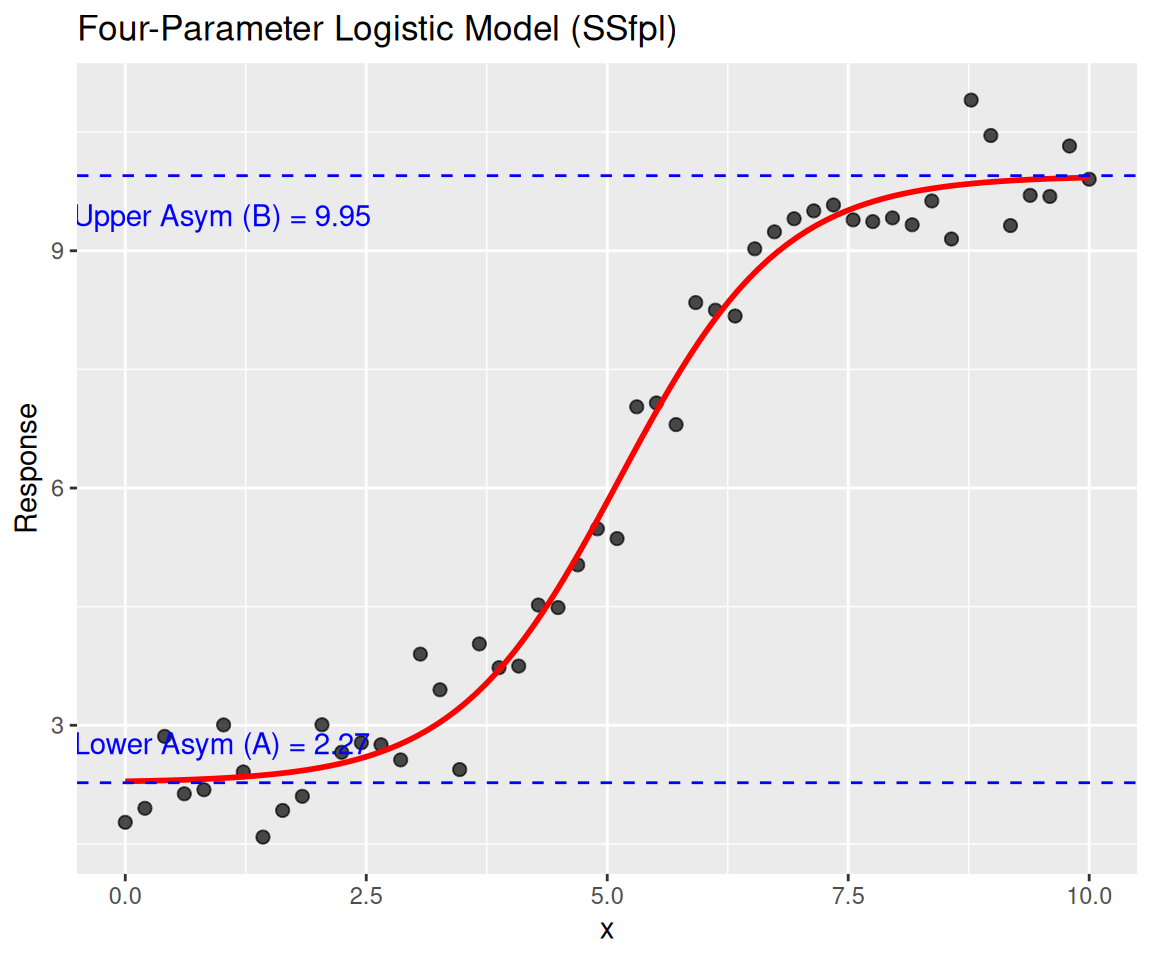
SSweibullModel:
\(y = Asym - (Asym - Drop) e^{-exp((x - lrc) / pwr)}\)
Parameters:
Asym: Asymptote.Drop: Drop parameter.lrc: Natural logarithm of the rate constant.pwr: Power parameter.# Set seed for reproducibility
set.seed(123)
# Simulate Weibull growth data
x <- 1:30
y <- SSweibull(x, Asym = 100, Drop = 80, lrc = log(0.1), pwr = 2) + rnorm(30, sd = 3)
df <- data.frame(x, y)
# Fit Weibull model using self-starting function
weibull_fit <- nls(y ~ SSweibull(x, Asym, Drop, lrc, pwr), data = df)
# Model summary and parameters
summary(weibull_fit)
Formula: y ~ SSweibull(x, Asym, Drop, lrc, pwr)
Parameters:
Estimate Std. Error t value Pr(>|t|)
Asym 99.5733 0.6026 165.249 < 2e-16 ***
Drop 82.0570 6.3274 12.969 7.36e-13 ***
lrc -2.2490 0.4594 -4.895 4.43e-05 ***
pwr 2.0591 0.3370 6.110 1.85e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.964 on 26 degrees of freedom
Number of iterations to convergence: 0
Achieved convergence tolerance: 8.536e-07
Key Parameters:
Upper Asymptote (Asym): 99.57331
Lower Asymptote: 17.51629
Rate Constant (exp(lrc)): 0.1055008
Power Parameter: 2.059132# Generate predictions for smooth curve
new_data <- data.frame(x = seq(1, 30, length.out = 300))
predicted <- predict(weibull_fit, newdata = new_data)
# Create visualization
ggplot(df, aes(x = x, y = y)) +
geom_point(size = 2.5, alpha = 0.7, color = "darkblue") +
geom_line(data = data.frame(x = new_data$x, y = predicted),
color = "red", linewidth = 1.2) +
geom_hline(yintercept = coef(weibull_fit)["Asym"],
linetype = "dashed", color = "darkgreen") +
geom_hline(yintercept = coef(weibull_fit)["Asym"] - coef(weibull_fit)["Drop"],
linetype = "dashed", color = "purple") +
annotate("text", x = 5, y = coef(weibull_fit)["Asym"] + 5,
label = paste("Upper Asymptote =", round(coef(weibull_fit)["Asym"], 1)),
color = "darkgreen") +
annotate("text", x = 5, y = (coef(weibull_fit)["Asym"] - coef(weibull_fit)["Drop"]) - 5,
label = paste("Lower Asymptote =",
round(coef(weibull_fit)["Asym"] - coef(weibull_fit)["Drop"], 1)),
color = "purple") +
labs(title = "Weibull Growth Curve Fit using SSweibull",
subtitle = "Red: Fitted Curve | Dashed: Upper/Lower Asymptotes",
x = "Time/Input Variable",
y = "Response") +
theme_minimal()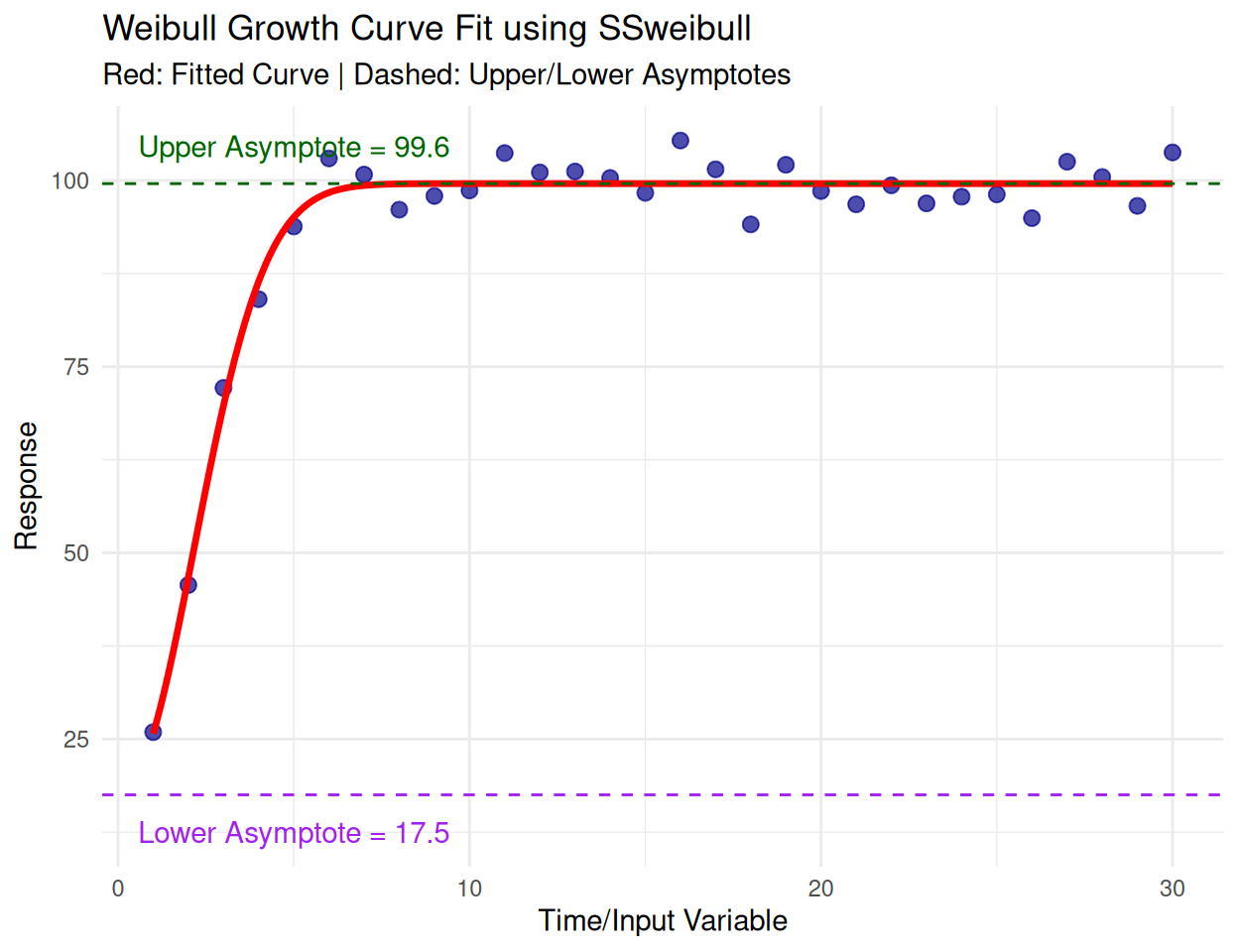
SSgompertzModel:
\(y = Asym \cdot e^{-b_2 \cdot e^{-b_3 \cdot x}}\)
Parameters:
Asym: Asymptote.b2: Displacement along the x-axis.b3: Growth rate.
Formula: y ~ SSgompertz(x, Asym, b2, b3)
Parameters:
Estimate Std. Error t value Pr(>|t|)
Asym 87.88811 20.69380 4.247 0.00381 **
b2 6.00060 1.85009 3.243 0.01419 *
b3 0.74290 0.06791 10.939 1.18e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 4.416 on 7 degrees of freedom
Number of iterations to convergence: 0
Achieved convergence tolerance: 1.909e-06# Extract fitted values
data$fitted <- predict(fit)
# Plot the data and the fitted model using ggplot2
ggplot(data, aes(x = x, y = y)) +
geom_point(color = 'blue', size = 2) +
geom_line(aes(y = fitted), color = 'red', size = 1) +
labs(title = "Gompertz Growth Model",
x = "X",
y = "Y") +
theme_minimal()Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.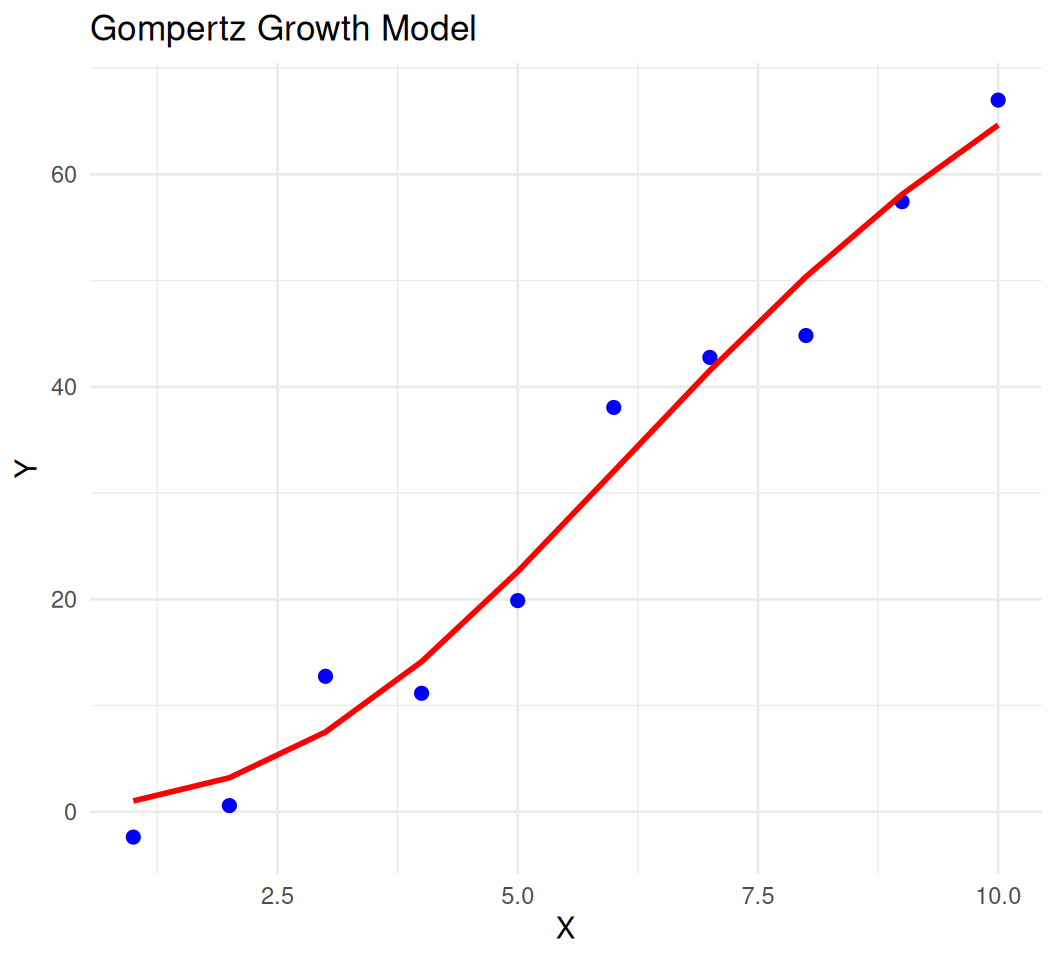
SSasympModel:
\(y = Asym + (R_0 - Asym) e^{-e^{lrc} \cdot x}\)
Parameters:
Asym: Horizontal asymptote.R0: y-intercept (at (x = 0)).
Formula: y ~ SSasymp(x, Asym, R0, lrc)
Parameters:
Estimate Std. Error t value Pr(>|t|)
Asym 48.4343 1.5058 32.16 <2e-16 ***
R0 100.0796 1.6400 61.02 <2e-16 ***
lrc -2.3718 0.0936 -25.34 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.97 on 27 degrees of freedom
Number of iterations to convergence: 0
Achieved convergence tolerance: 2.202e-07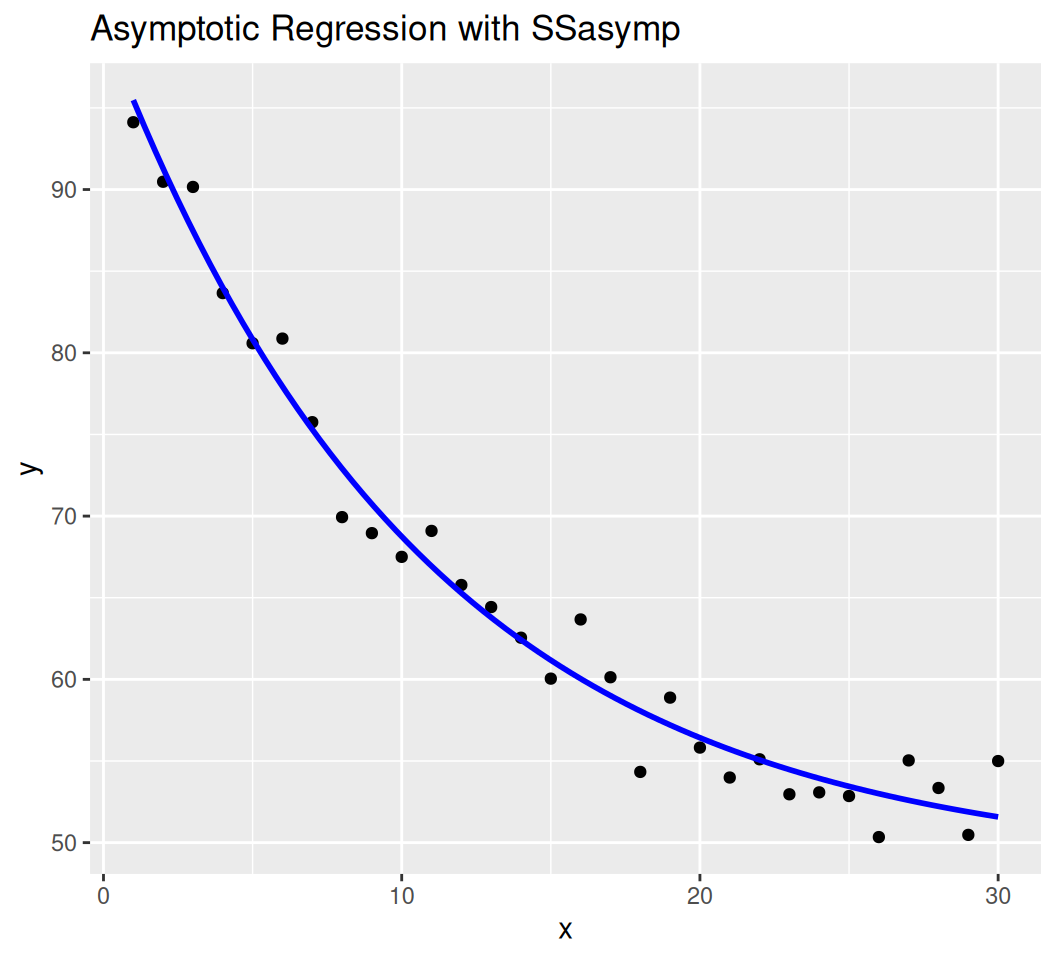
SSmicmen$y = \frac{V_m \cdot x}{K + x}$) Vm: Maximum reaction rate.K: Substrate concentration at half of \(V_m\).set.seed(123)
x <- seq(0, 50, by = 5)
y <- SSmicmen(x, Vm = 10, K = 5) + rnorm(length(x), sd = 0.5)
df <- data.frame(x, y)
# Get reasonable starting values
Vm_start <- max(df$y) * 1.1 # Slightly above observed max
K_start <- median(df$x[df$y >= 0.5 * max(df$y)]) # x near half-maximal response
# Fit model
fit <- nls(
y ~ SSmicmen(x, Vm, K),
data = df,
start = list(Vm = Vm_start, K = K_start),
control = nls.control(warnOnly = TRUE) # Show warnings instead of errors
)
# Check results
summary(fit)
Formula: y ~ SSmicmen(x, Vm, K)
Parameters:
Estimate Std. Error t value Pr(>|t|)
Vm 9.9403 0.3781 26.293 8.04e-10 ***
K 4.4053 0.9343 4.715 0.0011 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4946 on 9 degrees of freedom
Number of iterations to convergence: 6
Achieved convergence tolerance: 4.157e-06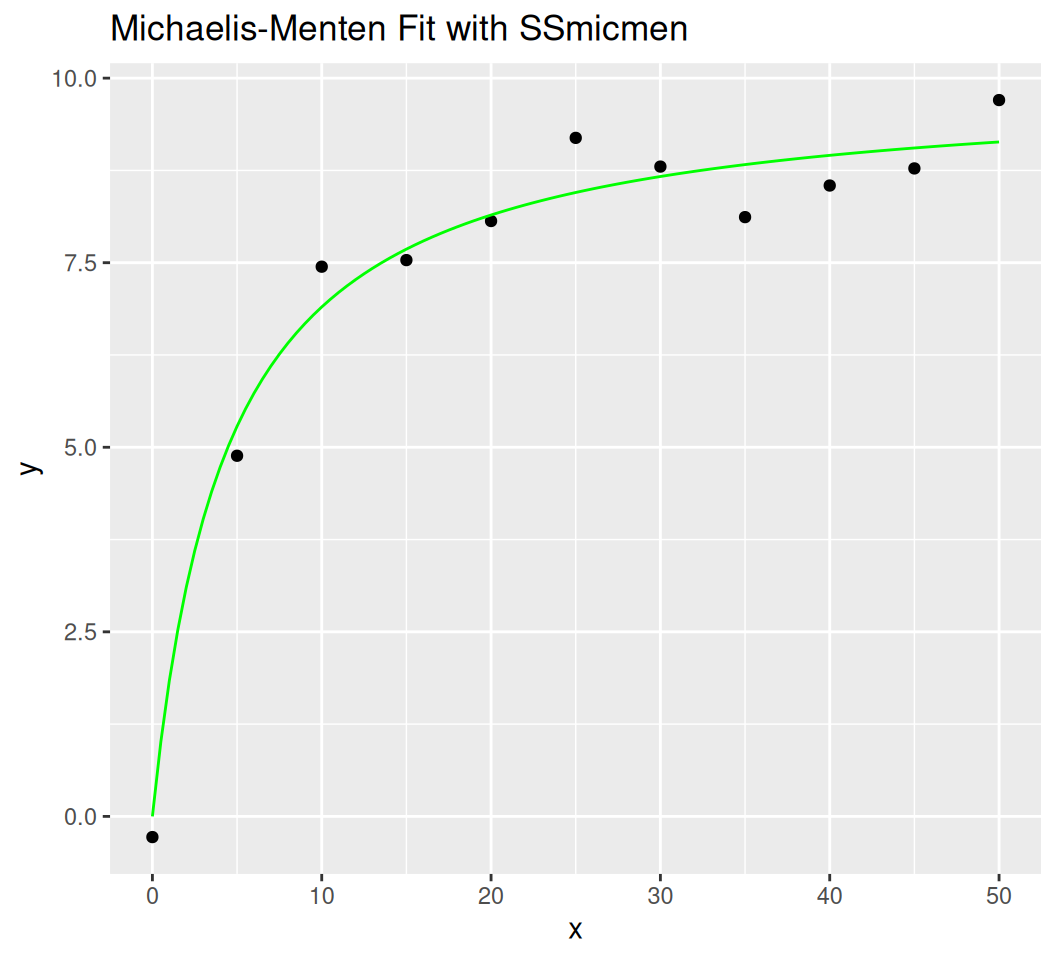
This tutorial explored the concept of self-starting functions in R, which are useful for fitting non-linear models. Self-starting functions provide initial parameter estimates that help the fitting algorithm converge more efficiently. We discussed several key self-starting functions, including the logistic model, four-parameter logistic model, Weibull growth curve, Gompertz growth model, asymptotic regression, and Michaelis-Menten model. For each function, we provided a brief description, example code, and visualization to demonstrate how to use them in practice. By leveraging self-starting functions, you can streamline the process of fitting non-linear models and obtain more reliable results.
The-R Book by Michael J. Crawley
A collection of self-starters for nonlinear regression in R - This article on R-bloggers discusses various self-starting functions available in R and how they can be used to simplify non-linear regression analysis.
Self-starting routines for nonlinear regression models - Another article on R-bloggers that delves into self-starting routines provided by the drc package, useful for dose-response analyses and other biological processes.
Some useful equations for nonlinear regression in R - This resource on StatForBiology provides useful equations and discusses the convenience of using self-starting routines in non-linear regression models.
Self-starting routines for nonlinear regression models - This article on StatForBiology explains the benefits of using self-starting functions in R for fitting non-linear models, especially in biological research.