Rows: 4,406
Columns: 13
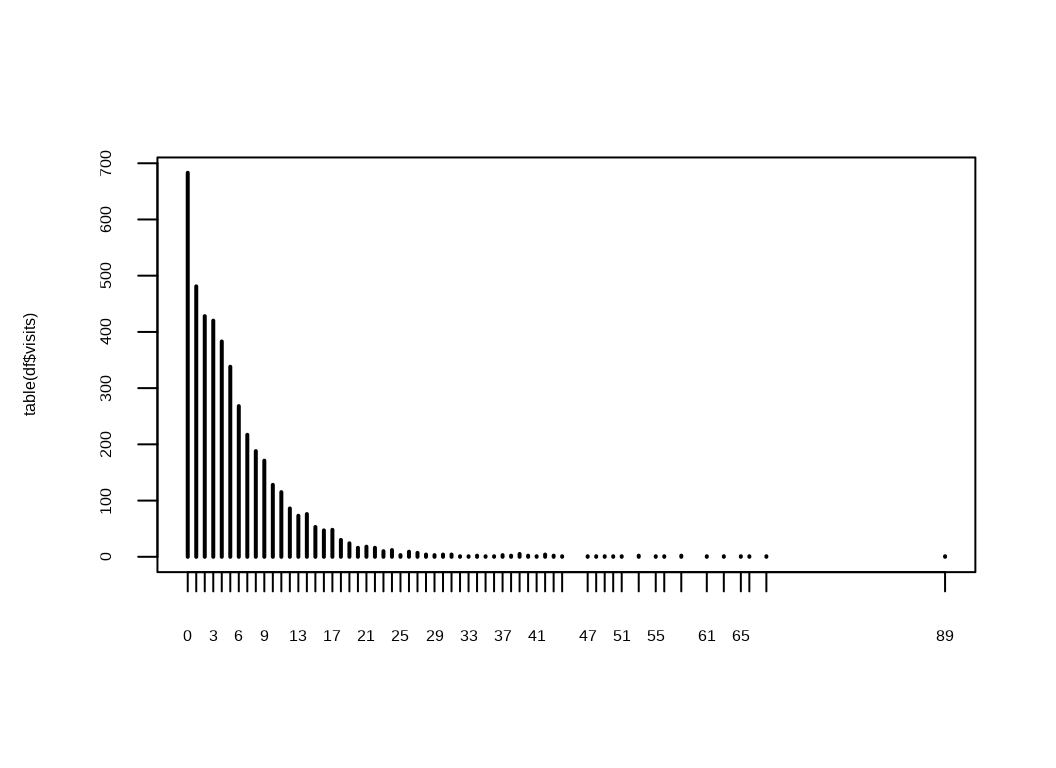
$ visits <int> 5, 1, 13, 16, 3, 17, 9, 3, 1, 0, 0, 44, 2, 1, 19, 19, 0, 3, …
$ hospital <int> 1, 0, 3, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, …
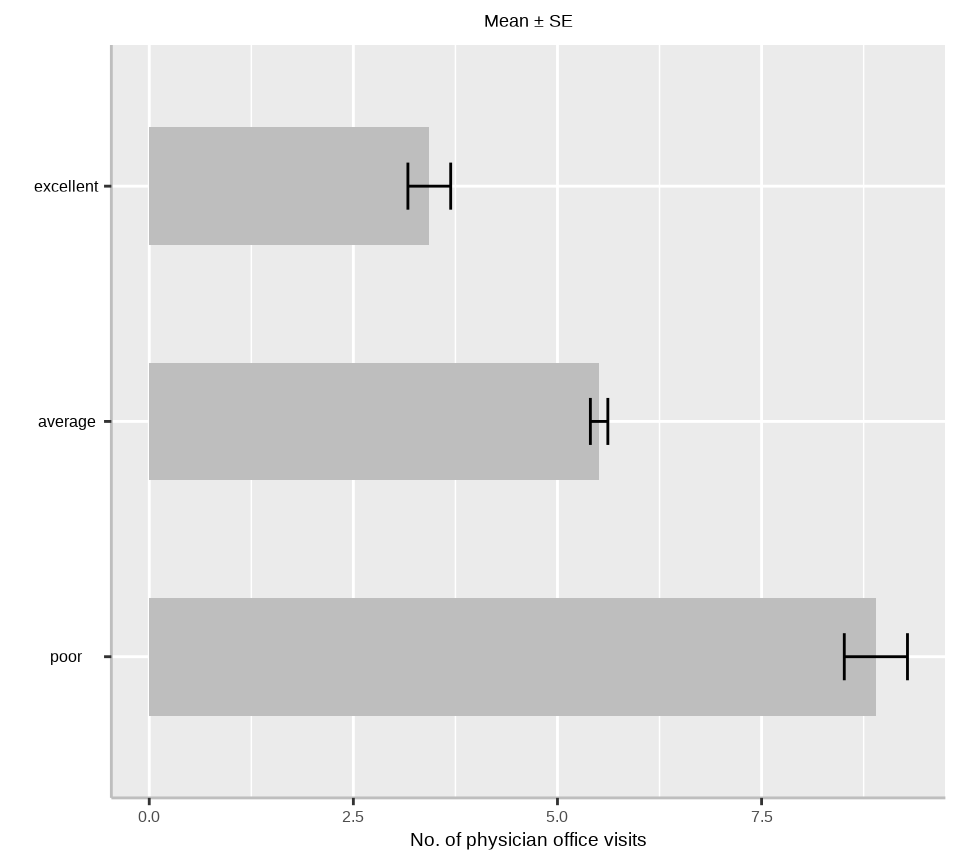
$ health <fct> average, average, poor, poor, average, poor, average, averag…
$ chronic <int> 2, 2, 4, 2, 2, 5, 0, 0, 0, 0, 1, 5, 1, 1, 1, 0, 1, 2, 3, 4, …
$ age <dbl> 6.9, 7.4, 6.6, 7.6, 7.9, 6.6, 7.5, 8.7, 7.3, 7.8, 6.6, 6.9, …
$ afam <fct> yes, no, yes, no, no, no, no, no, no, no, no, no, no, no, no…
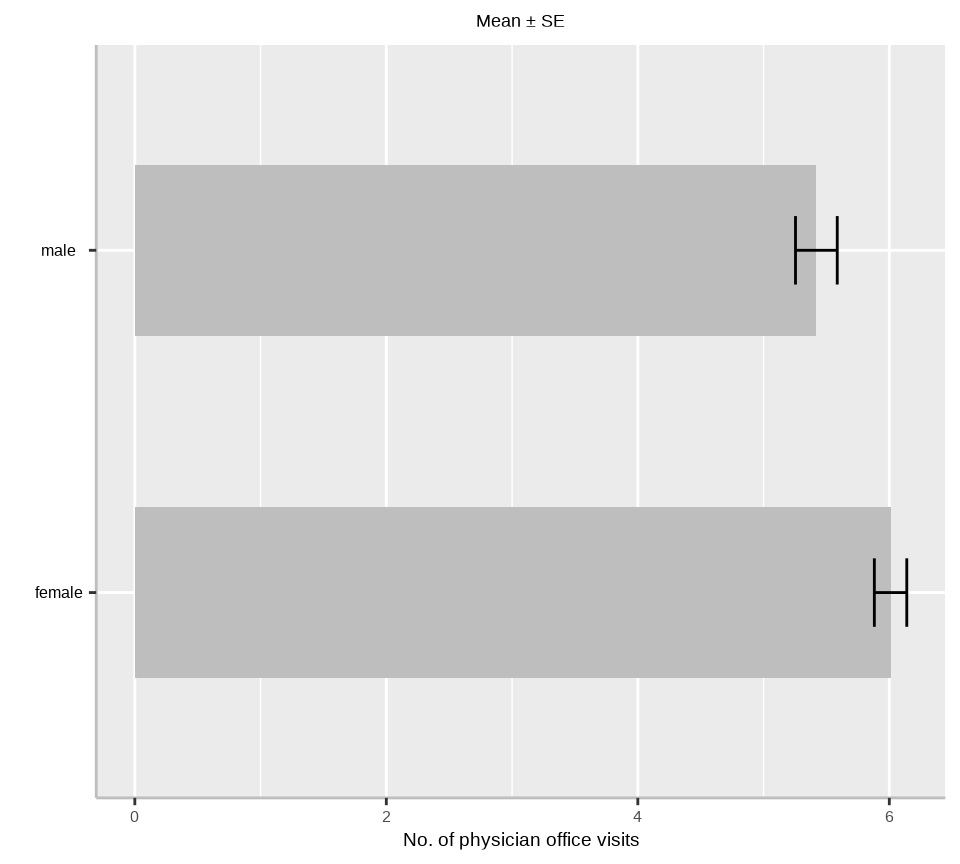
$ gender <fct> male, female, female, male, female, female, female, female, …
$ married <fct> yes, yes, no, yes, yes, no, no, no, no, no, yes, yes, no, no…
$ school <int> 6, 10, 10, 3, 6, 7, 8, 8, 8, 8, 8, 15, 8, 8, 12, 8, 8, 8, 10…
$ income <dbl> 2.881000, 2.747800, 0.653200, 0.658800, 0.658800, 0.330100, …
$ employed <fct> yes, no, no, no, no, no, no, no, no, no, yes, no, no, no, no…
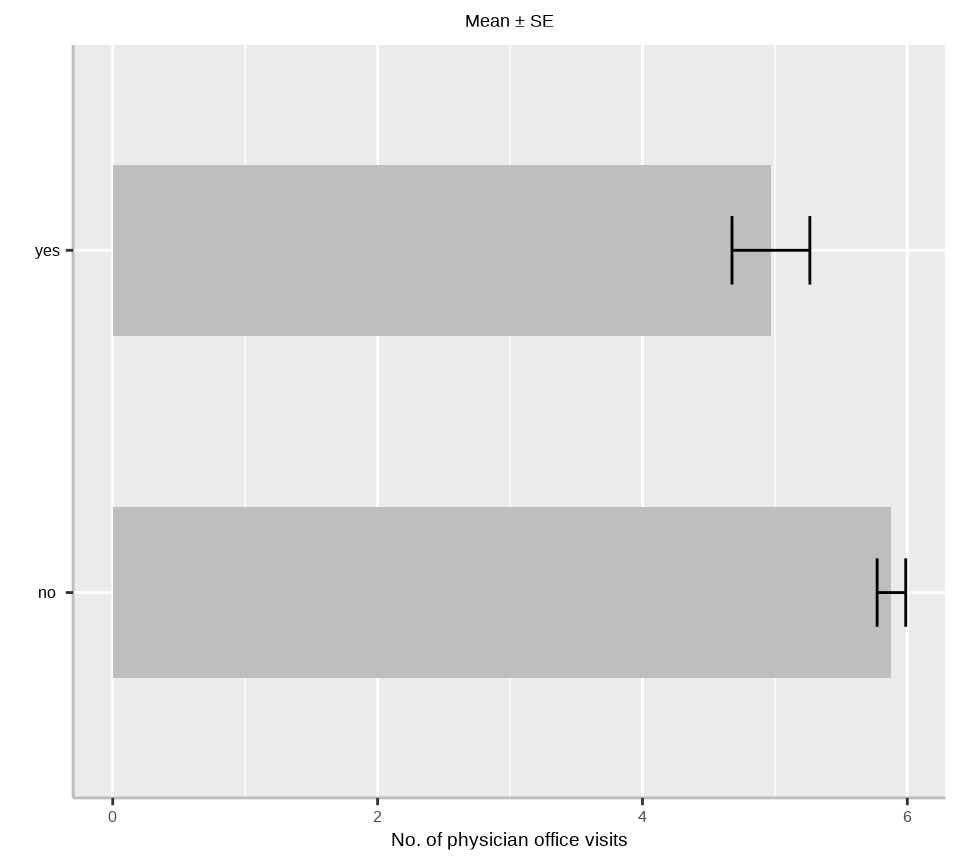
$ insurance <fct> yes, yes, no, yes, yes, no, yes, yes, yes, yes, yes, yes, no…
$ medicaid <fct> no, no, yes, no, no, yes, no, no, no, no, no, no, no, yes, n…