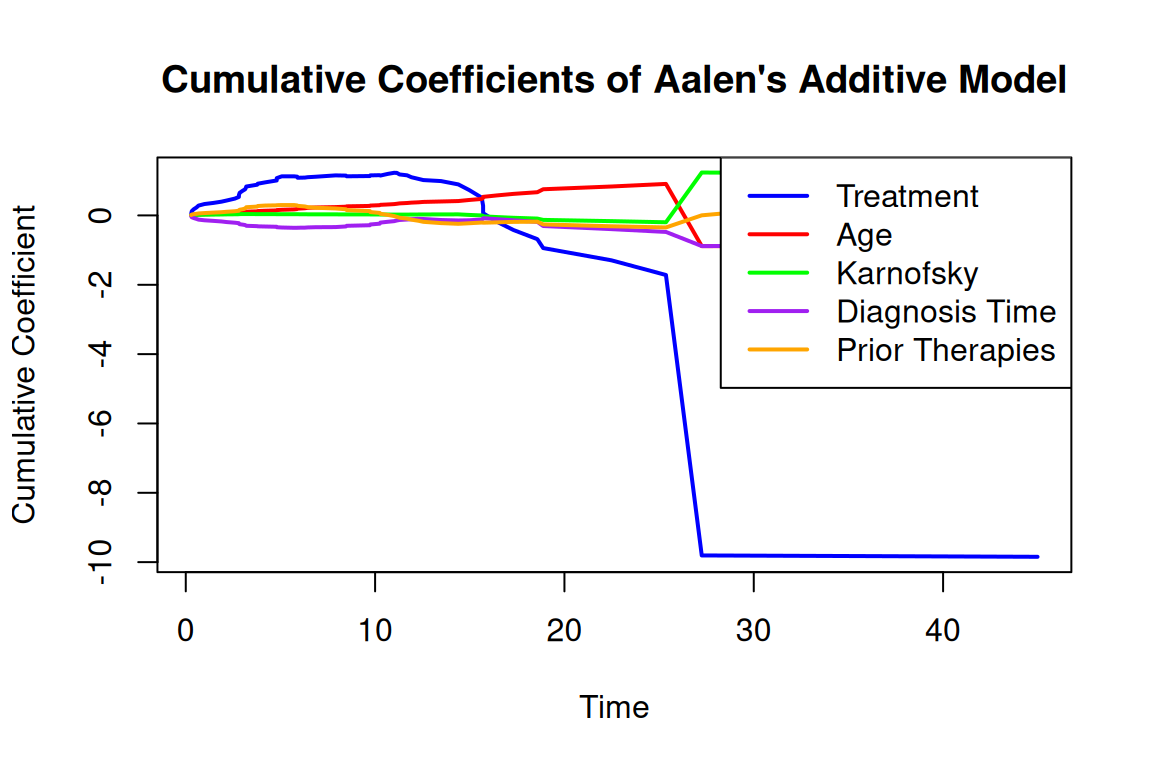
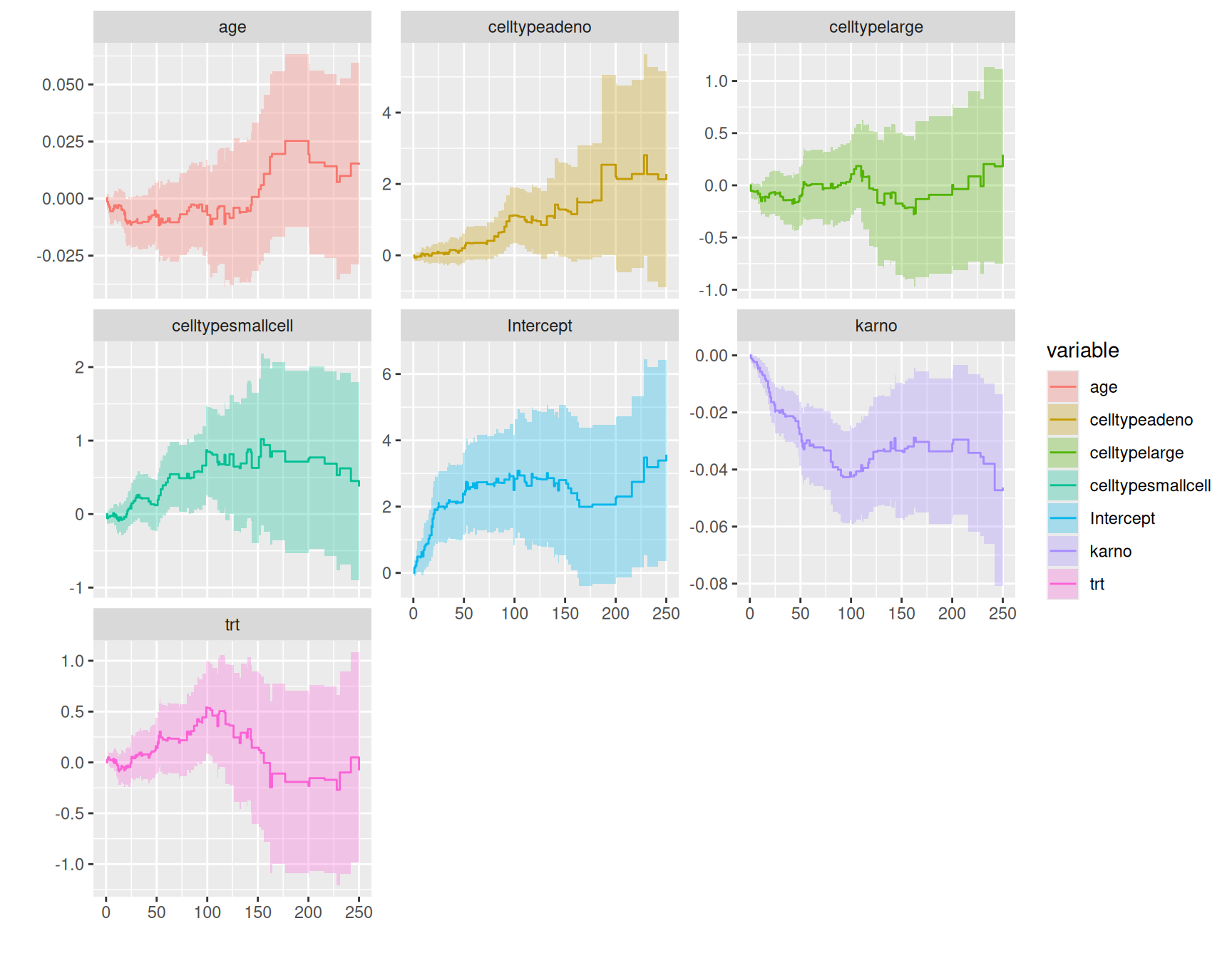
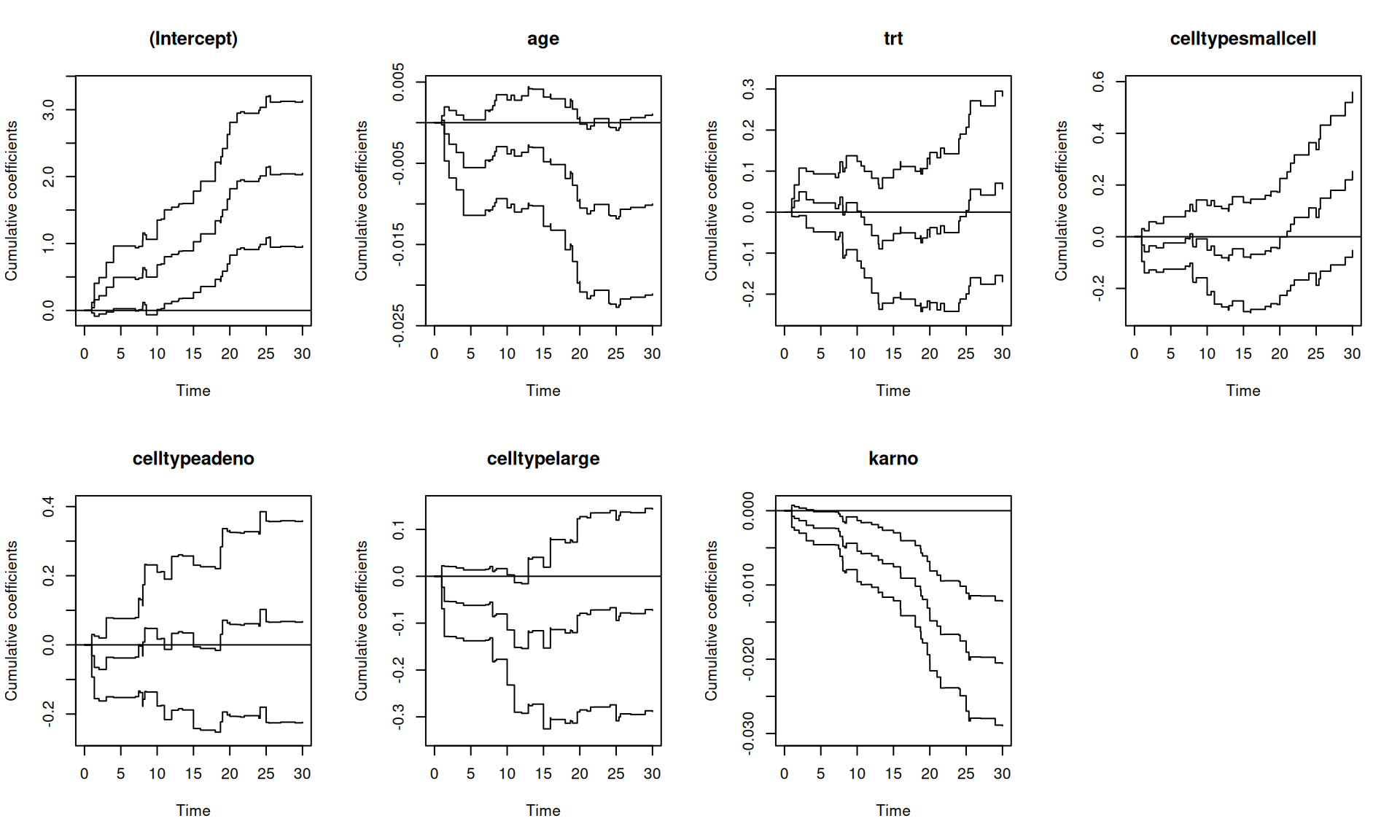
Rows: 137
Columns: 8
$ trt <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ celltype <fct> squamous, squamous, squamous, squamous, squamous, squamous, s…
$ time <dbl> 72, 411, 228, 126, 118, 10, 82, 110, 314, 100, 42, 8, 144, 25…
$ status <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0…
$ karno <dbl> 60, 70, 60, 60, 70, 20, 40, 80, 50, 70, 60, 40, 30, 80, 70, 6…
$ diagtime <dbl> 7, 5, 3, 9, 11, 5, 10, 29, 18, 6, 4, 58, 4, 9, 11, 3, 9, 2, 4…
$ age <dbl> 69, 64, 38, 63, 65, 49, 69, 68, 43, 70, 81, 63, 63, 52, 48, 6…
$ prior <dbl> 0, 10, 0, 10, 10, 0, 10, 0, 0, 0, 0, 10, 0, 10, 10, 0, 0, 0, …