Rows: 422
Columns: 16
$ SEASON <chr> "Rabi 12-13", "Rabi 12-13", "Rabi 12-13", "Rabi 12-13", …
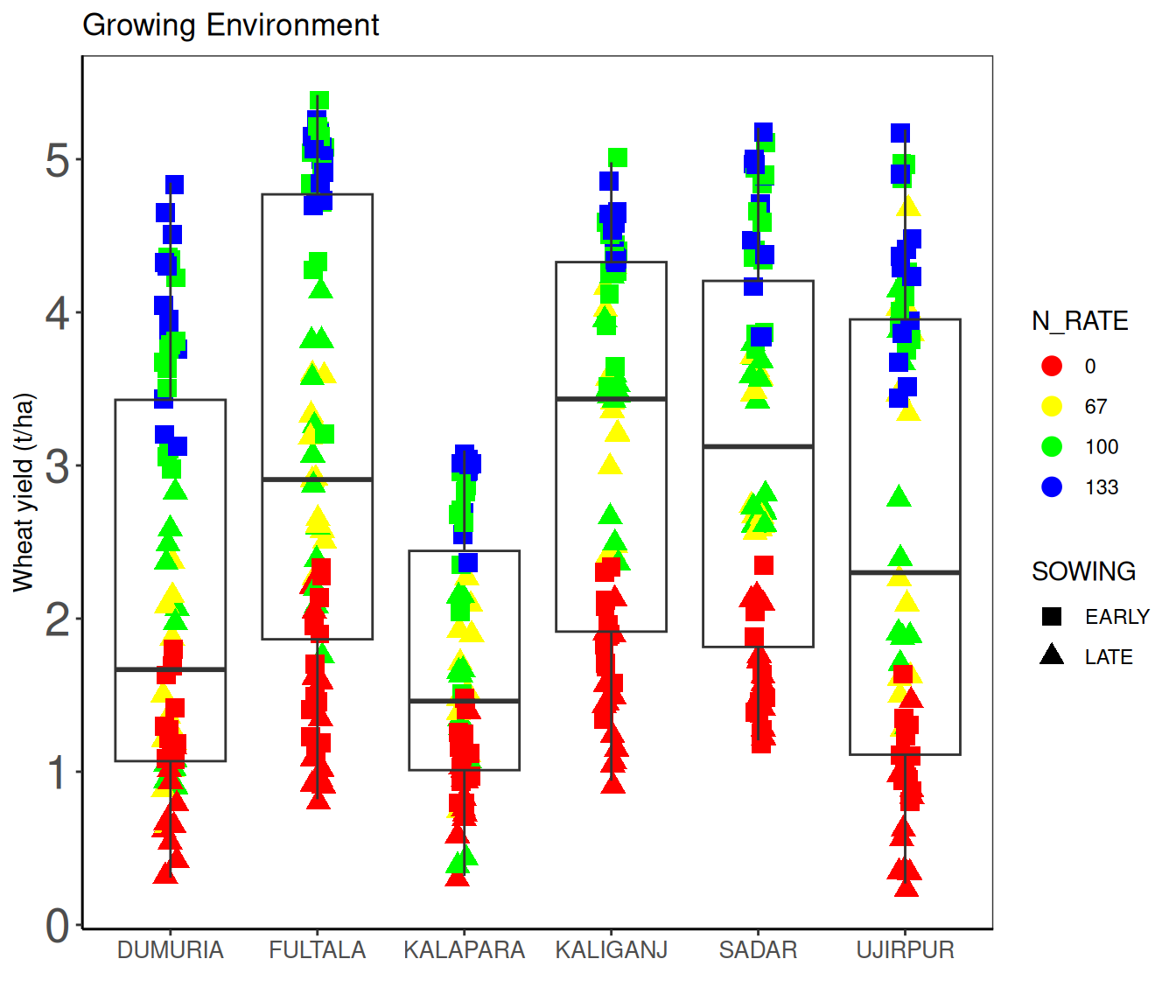
$ ENV <chr> "KALAPARA", "KALAPARA", "KALAPARA", "KALAPARA", "KALAPAR…
$ SOWING <chr> "LATE", "LATE", "LATE", "LATE", "LATE", "LATE", "LATE", …
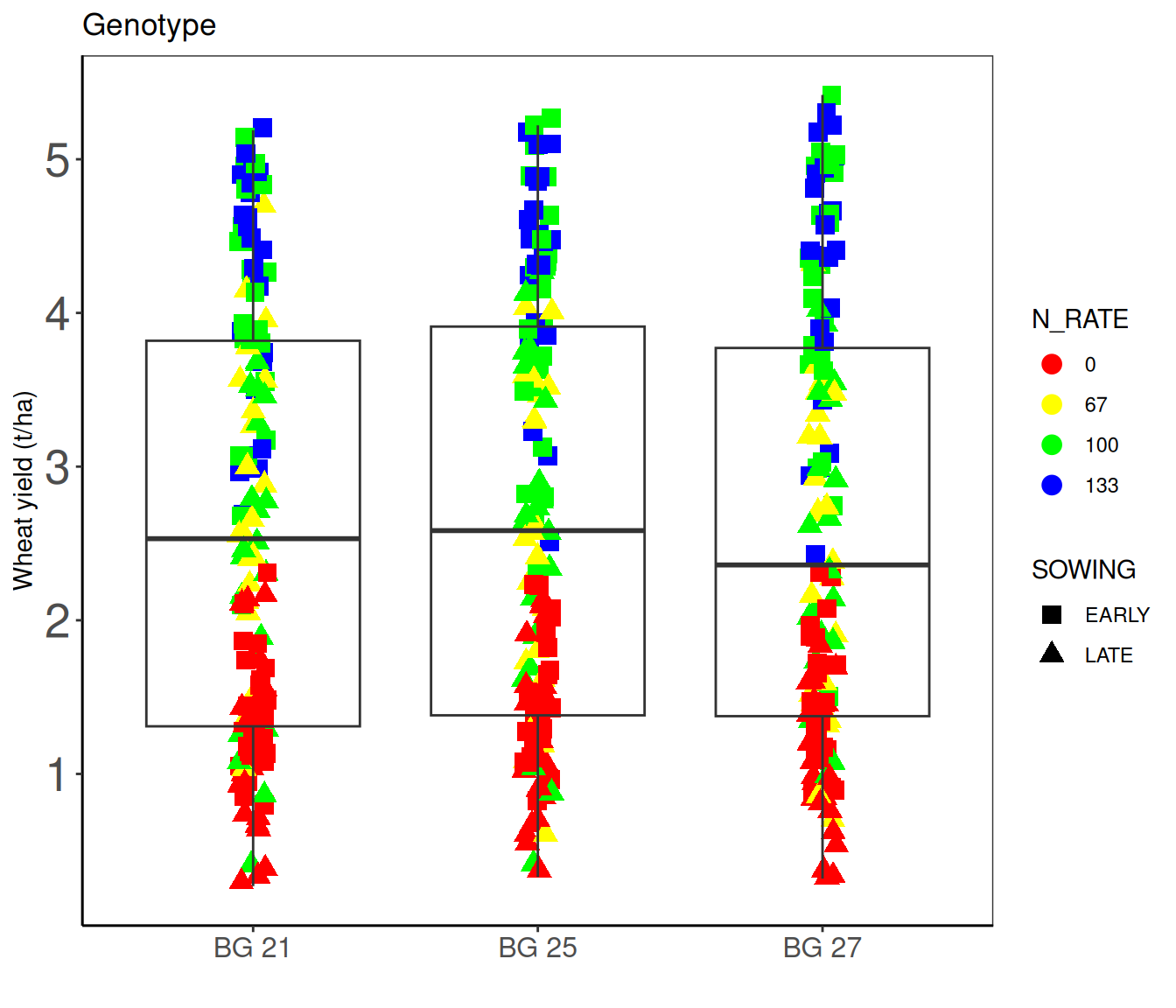
$ GEN <chr> "BG 27", "BG 25", "BG 21", "BG 27", "BG 25", "BG 21", "B…
$ N_RATE <dbl> 0, 0, 0, 67, 67, 67, 100, 100, 100, 0, 0, 67, 67, 67, 10…
$ DATE_SOW <chr> "1/5/2013", "1/5/2013", "1/5/2013", "1/5/2013", "1/5/201…
$ Optimum_date <chr> "12/15/2012", "12/15/2012", "12/15/2012", "12/15/2012", …
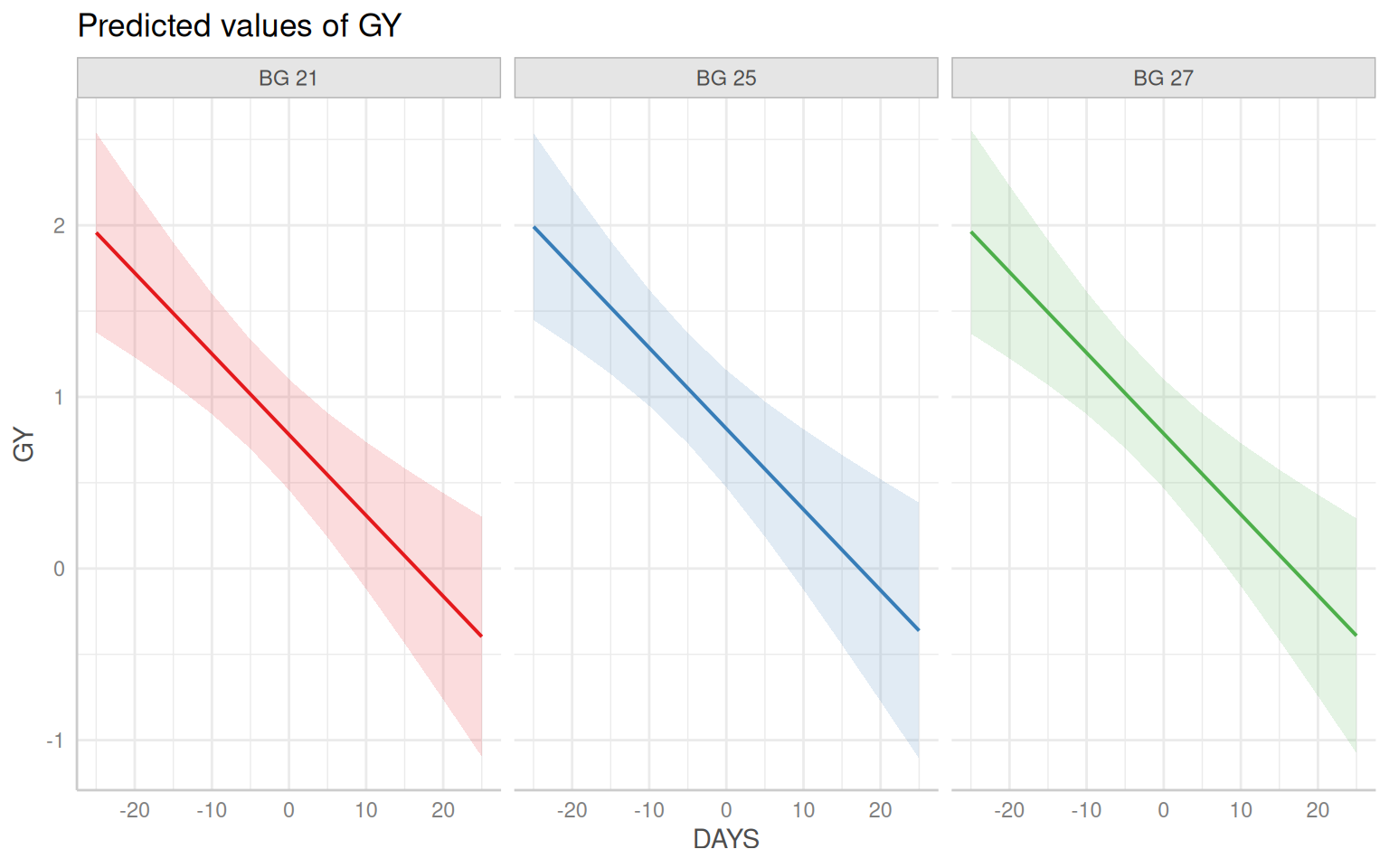
$ DAYS <dbl> 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, …
$ GY <dbl> 0.319, 0.622, 0.737, 0.732, 1.095, 1.348, 1.365, 0.407, …
$ STY <chr> "0.5699", "1.3067", "0.987", "1.2628", "0.251883333", "2…
$ Maturity_days <dbl> 103, 101, 102, 104, 105, 105, 104, 104, 105, 92, 92, 92,…
$ Weeding <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Weeding_No <chr> "1 weeding", "1 weeding", "1 weeding", "1 weeding", "1 w…
$ Irrigation <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1,…
$ Irrigation_No <chr> "2 irrigation", "2 irrigation", "2 irrigation", "2 irrig…
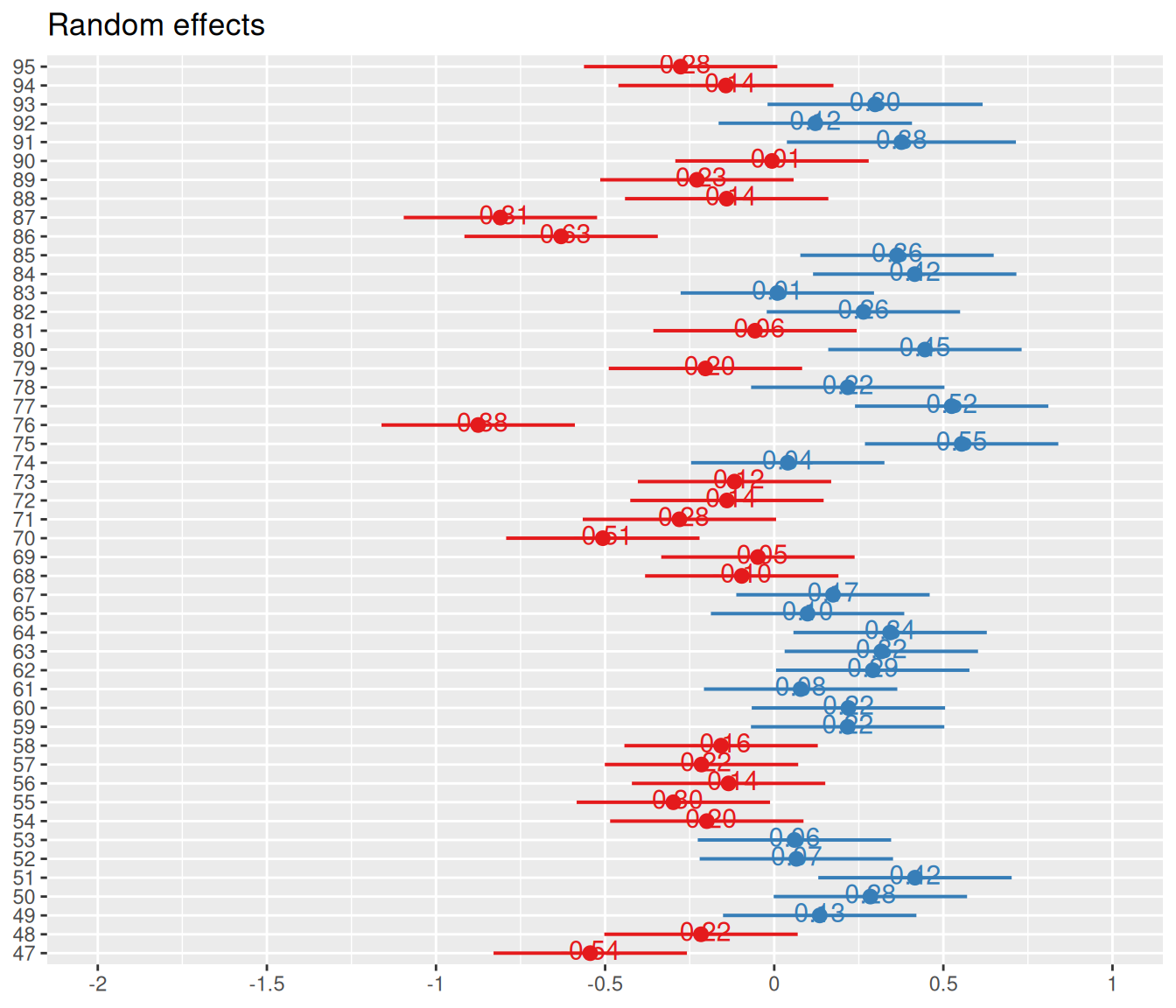
$ FARMER <dbl> 95, 95, 95, 95, 95, 95, 95, 95, 95, 94, 94, 94, 94, 94, …