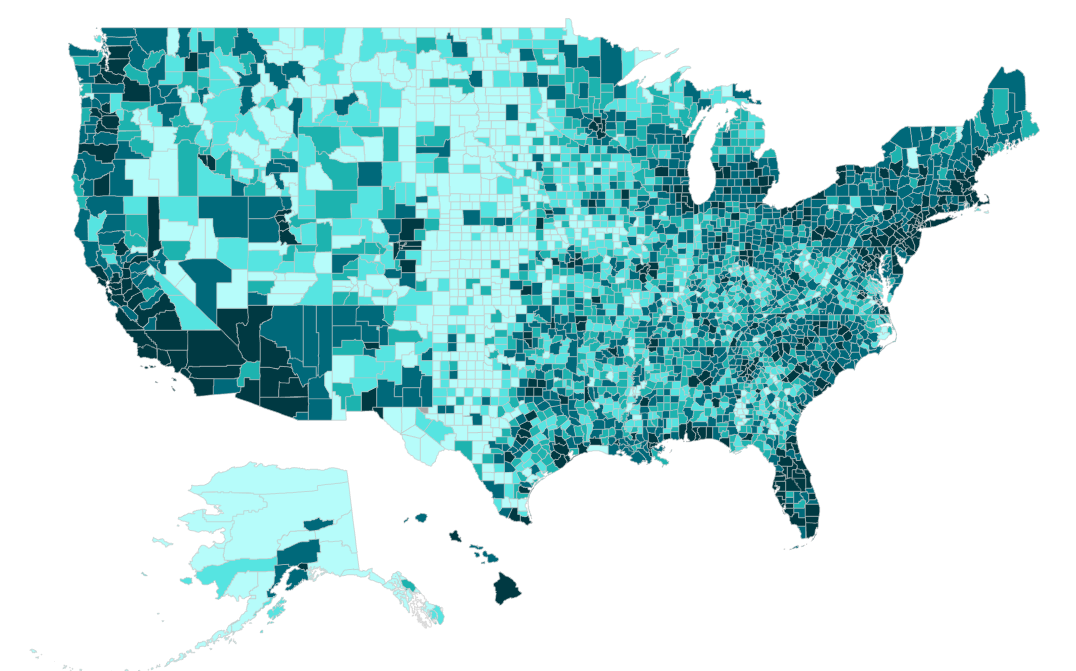
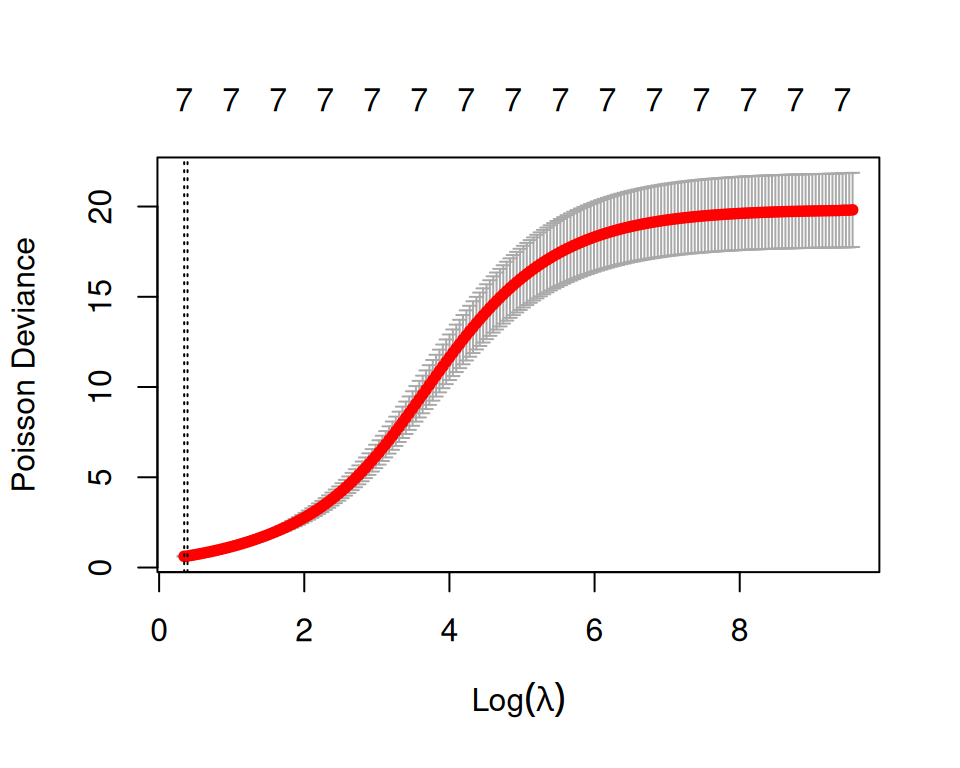
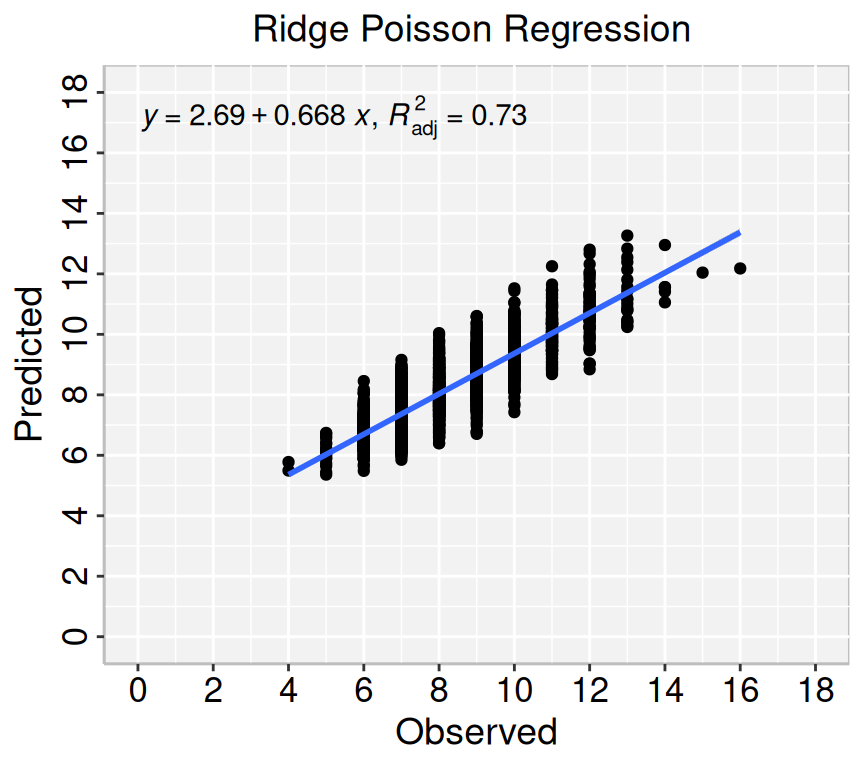
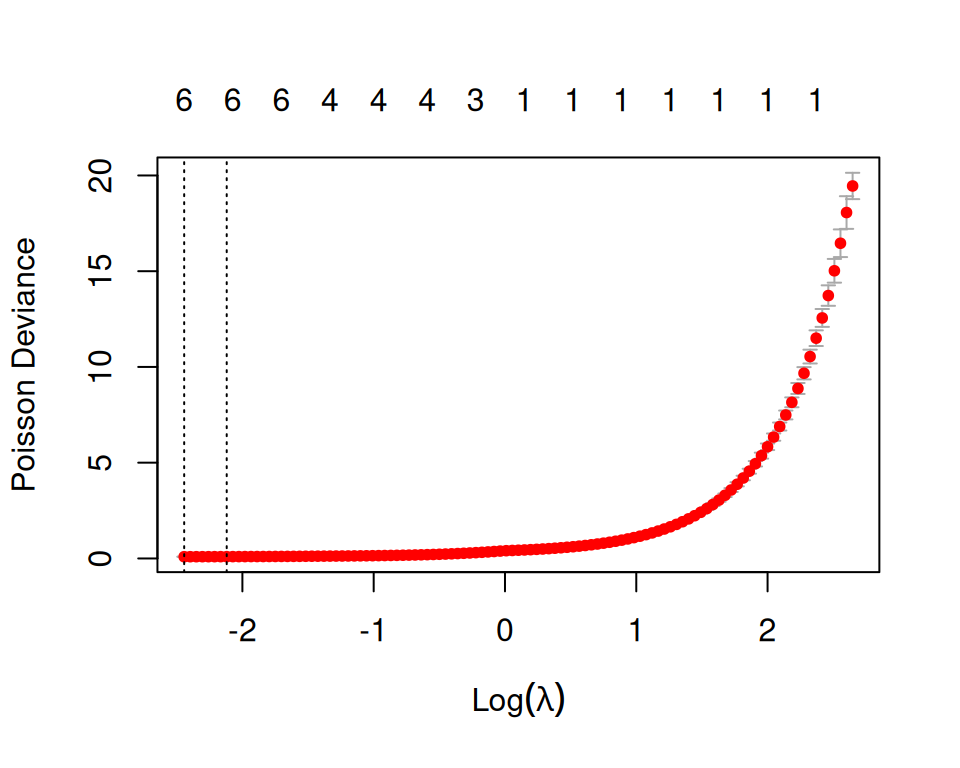
Rows: 3107 Columns: 15
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (3): State, County, Urban_Rural
dbl (12): FIPS, X, Y, POP_Total, Diabetes_count, Diabetes_per, Obesity, Acce...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Rows: 3,107
Columns: 8
$ Diabetes_per <dbl> 9.24, 8.48, 11.72, 10.08, 10.26, 9.06, 11.80, 13.2…
$ Obesity <dbl> 29.22, 28.94, 29.34, 29.44, 30.10, 19.86, 30.38, 3…
$ Physical_Inactivity <dbl> 26.42, 22.86, 23.72, 25.38, 24.76, 18.58, 28.66, 2…
$ Access_Excercise <dbl> 70.8, 72.2, 49.8, 30.6, 24.6, 19.6, 48.0, 51.4, 62…
$ Food_Env_Index <dbl> 6.9, 7.7, 5.5, 7.6, 8.1, 4.3, 6.5, 6.3, 6.4, 7.7, …
$ SVI <dbl> 0.5130, 0.3103, 0.9927, 0.8078, 0.5137, 0.8310, 0.…
$ Urban_Rural <chr> "Urban", "Urban", "Rural", "Urban", "Urban", "Rura…
$ POP_Total <dbl> 55707.0, 218346.8, 25078.2, 22448.2, 57852.4, 1017…