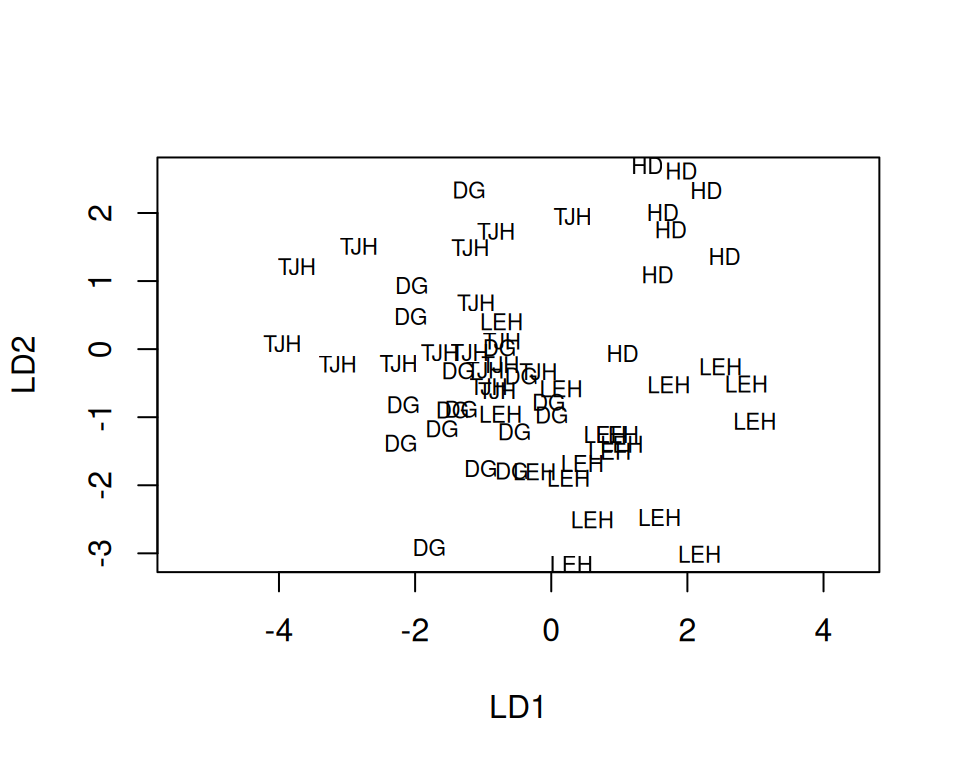
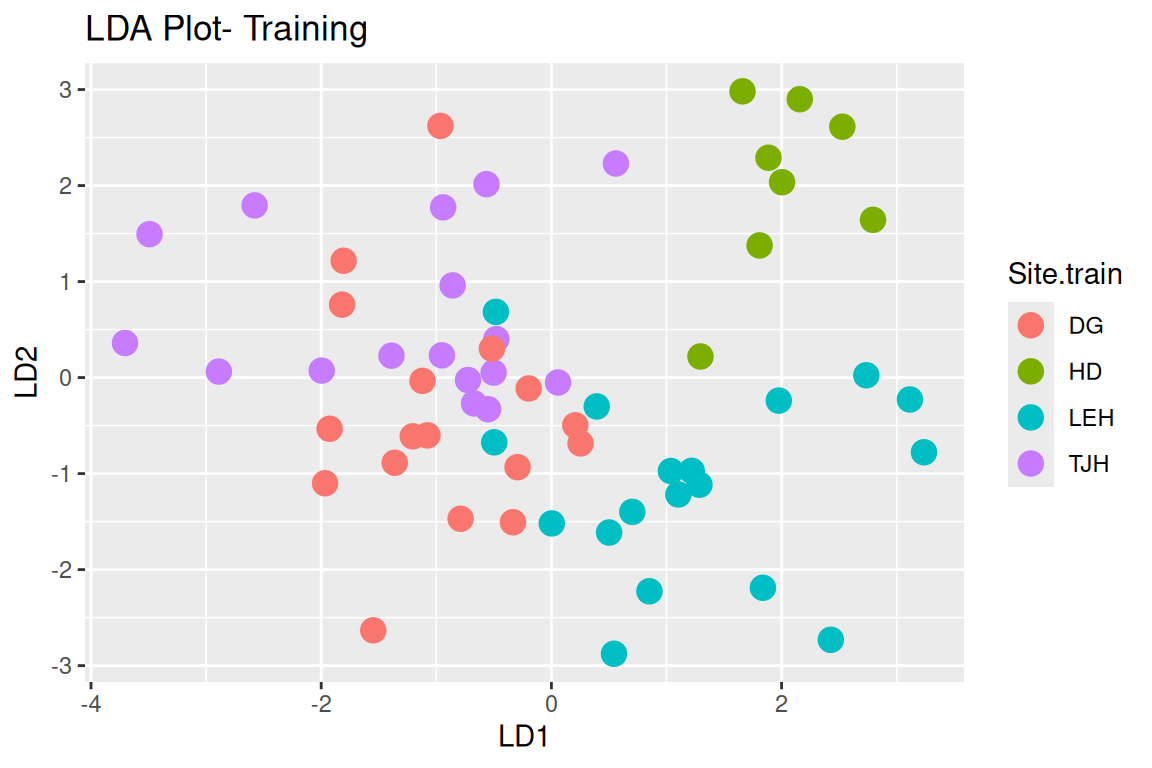
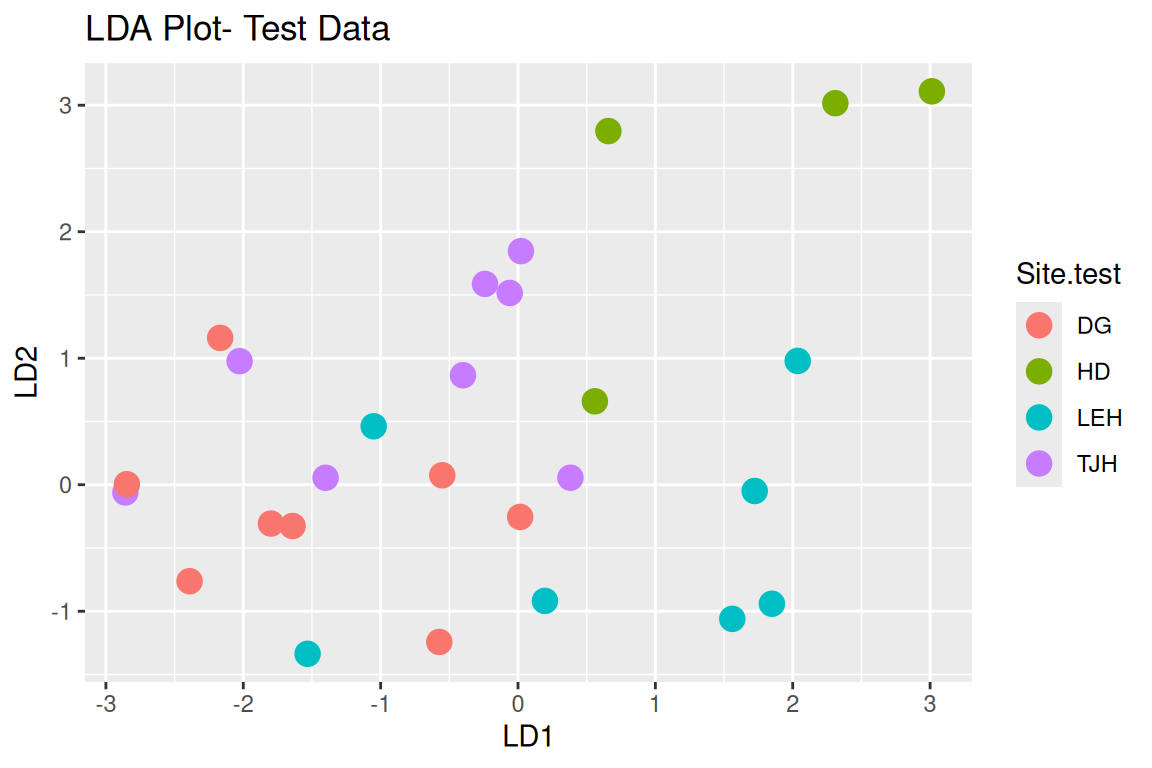
Rows: 87
Columns: 12
$ site <chr> "TJH", "TJH", "TJH", "TJH", "TJH", "TJH", "TJH", "TJH", "…
$ plant <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17…
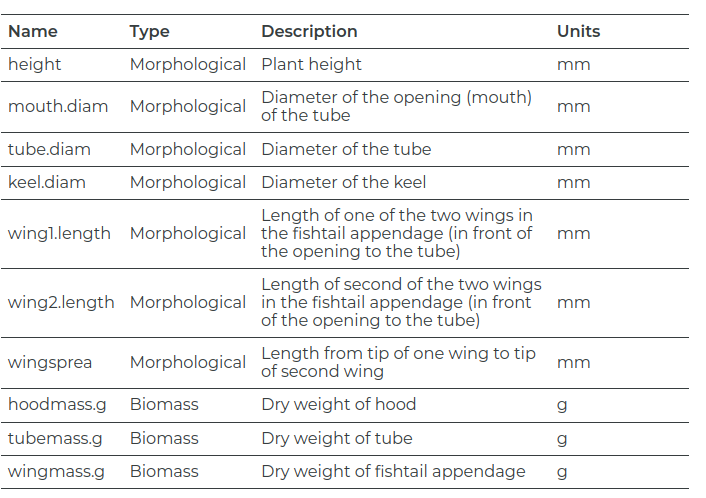
$ height <dbl> 654, 413, 610, 546, 665, 665, 638, 629, 508, 709, 468, 62…
$ mouth.diam <dbl> 38.4, 22.2, 31.2, 34.4, 30.5, 33.6, 37.4, 29.6, 22.9, 39.…
$ tube.diam <dbl> 16.6, 17.2, 19.9, 20.8, 20.4, 19.5, 23.0, 19.9, 20.4, 19.…
$ keel.diam <dbl> 6.4, 5.9, 6.7, 6.3, 6.6, 6.6, 7.4, 5.9, 8.2, 5.9, 7.7, 5.…
$ wing1.length <dbl> 85, 55, 62, 84, 60, 84, 63, 75, 52, 55, 56, 93, 90, 67, 4…
$ wing2.length <dbl> 76, 26, 60, 79, 51, 66, 70, 73, 53, 53, 16, 104, 96, 63, …
$ wingsprea <dbl> 55, 60, 78, 95, 30, 82, 86, 130, 75, 65, 60, 120, 145, 62…
$ hoodmass.g <dbl> 1.38, 0.49, 0.60, 1.12, 0.67, 1.27, 1.55, 0.83, 0.57, 1.3…
$ tubemass.g <dbl> 3.54, 1.48, 2.20, 2.95, 3.36, 4.05, 4.27, 3.48, 2.20, 4.4…
$ wingmass.g <dbl> 0.29, 0.06, 0.16, 0.24, 0.08, 0.21, 0.21, 0.19, 0.08, 0.1…