Rows: 3,443
Columns: 9
$ year <int> 1999, 1999, 1999, 1999, 1999, 1999, 1999, 1999, 1999, 1999, 1999…
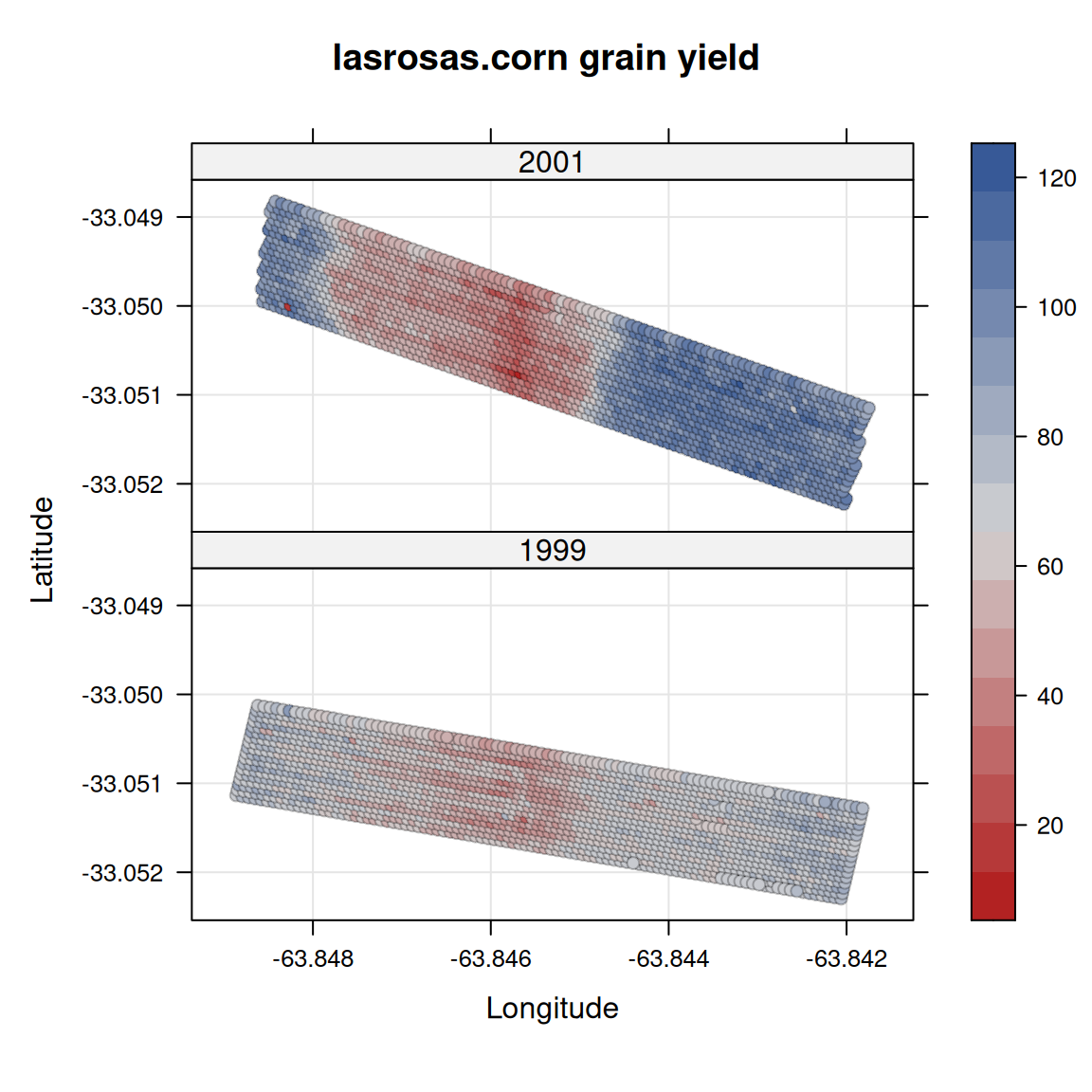
$ lat <dbl> -33.05113, -33.05115, -33.05116, -33.05117, -33.05118, -33.05120…
$ long <dbl> -63.84886, -63.84879, -63.84872, -63.84865, -63.84858, -63.84851…
$ yield <dbl> 72.14, 73.79, 77.25, 76.35, 75.55, 70.24, 76.17, 69.17, 69.77, 6…
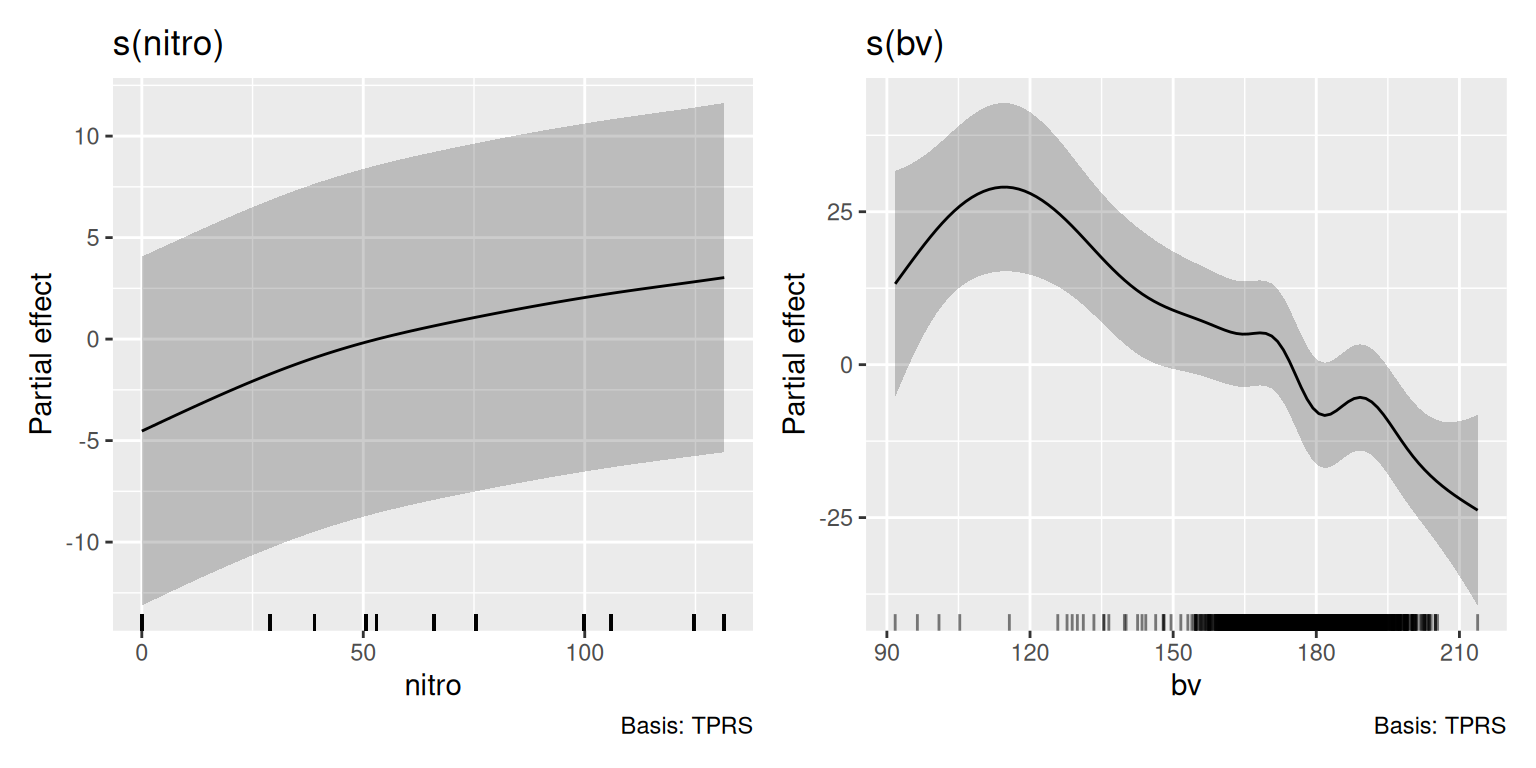
$ nitro <dbl> 131.5, 131.5, 131.5, 131.5, 131.5, 131.5, 131.5, 131.5, 131.5, 1…
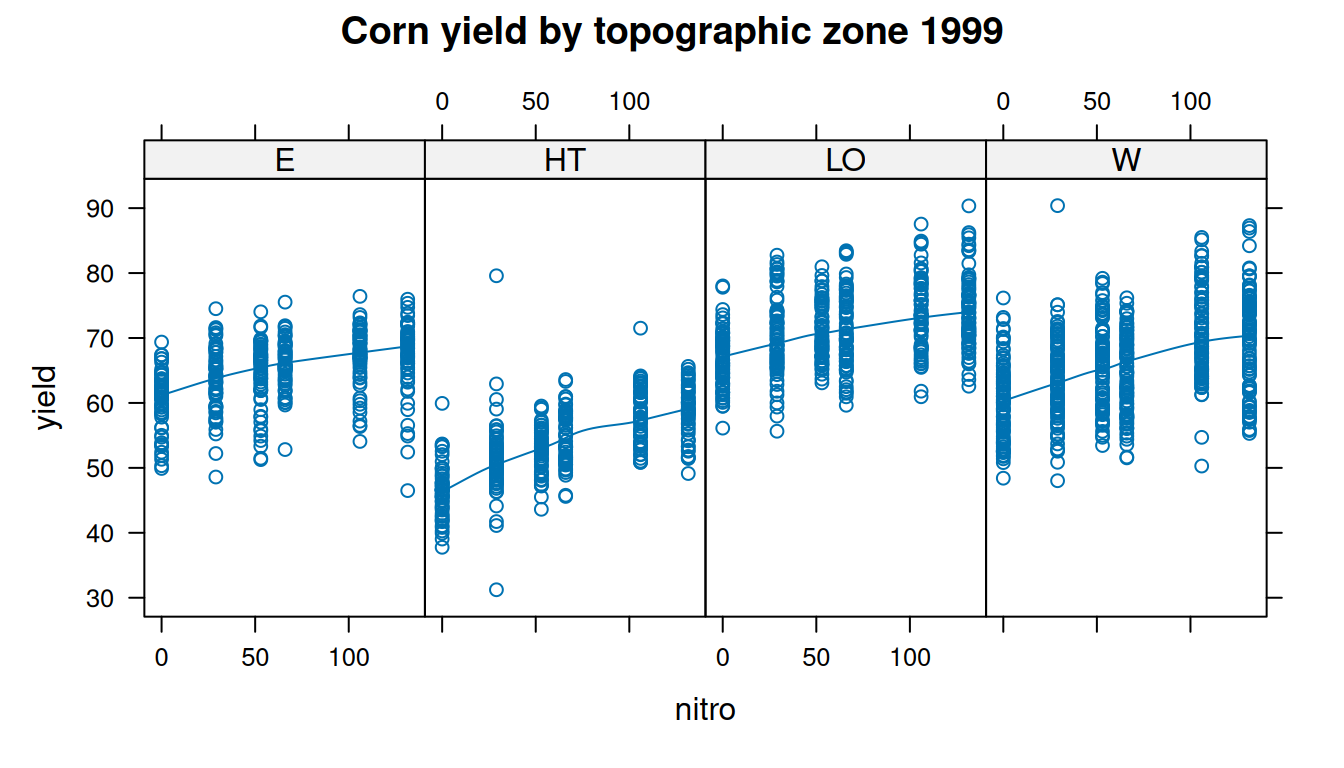
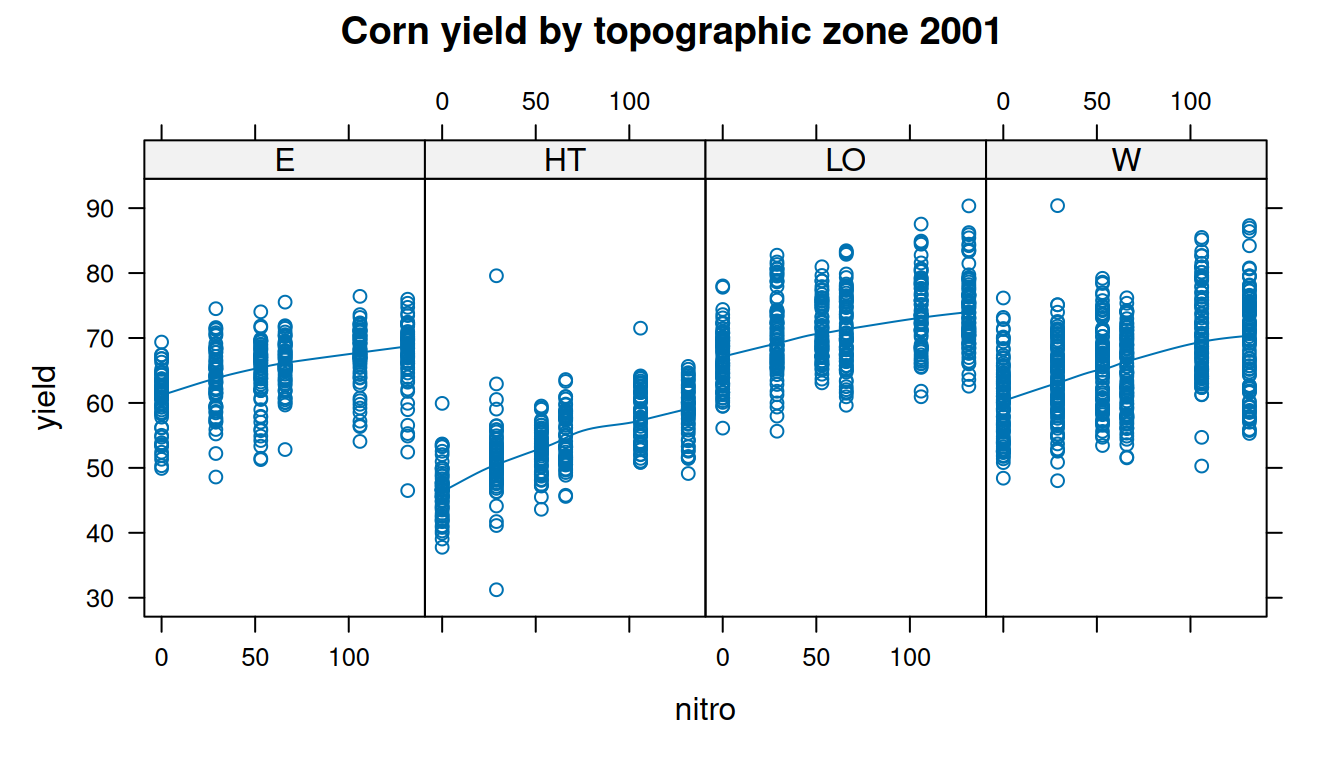
$ topo <fct> W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W, W…
$ bv <dbl> 162.60, 170.49, 168.39, 176.68, 171.46, 170.56, 172.94, 171.86, …
$ rep <fct> R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, R1, …
$ nf <fct> N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, N5, …